a83e4875099213b9d4a2f704438952f3.ppt
- Количество слайдов: 69
 Yeast as a model organism • Model eukaryote – Experimental genetics – Gene function – Orthologs, family members – Pathway function - “Biological synteny” • Testbed for genomic technologies – Genome sequenced (4/96) relatively less complex – Ability to assess biological relevance of the data
Yeast as a model organism • Model eukaryote – Experimental genetics – Gene function – Orthologs, family members – Pathway function - “Biological synteny” • Testbed for genomic technologies – Genome sequenced (4/96) relatively less complex – Ability to assess biological relevance of the data


 Genomics technology development Yeast as a testbed • Gene expression patterns – DNA microarrays, SAGE • Genomic DNA scans – Mapping complex traits (SNPs) • Phenotype screening – Genome-wide knockouts • Genetic interaction networks – Synthetic lethals • Protein interaction networks – Two-hybrid, mass spectrometry
Genomics technology development Yeast as a testbed • Gene expression patterns – DNA microarrays, SAGE • Genomic DNA scans – Mapping complex traits (SNPs) • Phenotype screening – Genome-wide knockouts • Genetic interaction networks – Synthetic lethals • Protein interaction networks – Two-hybrid, mass spectrometry
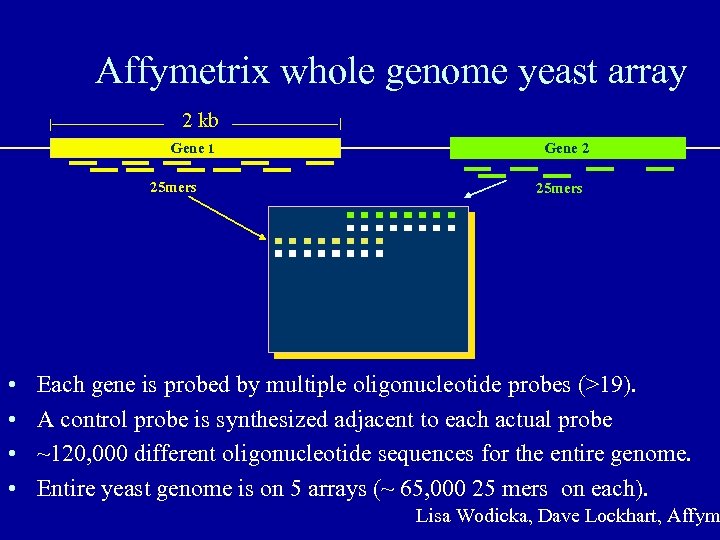 Affymetrix whole genome yeast array 2 kb Gene 1 25 mers • • Gene 2 25 mers Each gene is probed by multiple oligonucleotide probes (>19). A control probe is synthesized adjacent to each actual probe ~120, 000 different oligonucleotide sequences for the entire genome. Entire yeast genome is on 5 arrays (~ 65, 000 25 mers on each). Lisa Wodicka, Dave Lockhart, Affym
Affymetrix whole genome yeast array 2 kb Gene 1 25 mers • • Gene 2 25 mers Each gene is probed by multiple oligonucleotide probes (>19). A control probe is synthesized adjacent to each actual probe ~120, 000 different oligonucleotide sequences for the entire genome. Entire yeast genome is on 5 arrays (~ 65, 000 25 mers on each). Lisa Wodicka, Dave Lockhart, Affym
 Assigning function by analyzing gene expression • • • Isolate m. RNA Label m. RNA Hybridize to array Detect hybridization Measure the abundance of every m. RNA – Test different growth conditions – Test different genetic backgrounds
Assigning function by analyzing gene expression • • • Isolate m. RNA Label m. RNA Hybridize to array Detect hybridization Measure the abundance of every m. RNA – Test different growth conditions – Test different genetic backgrounds
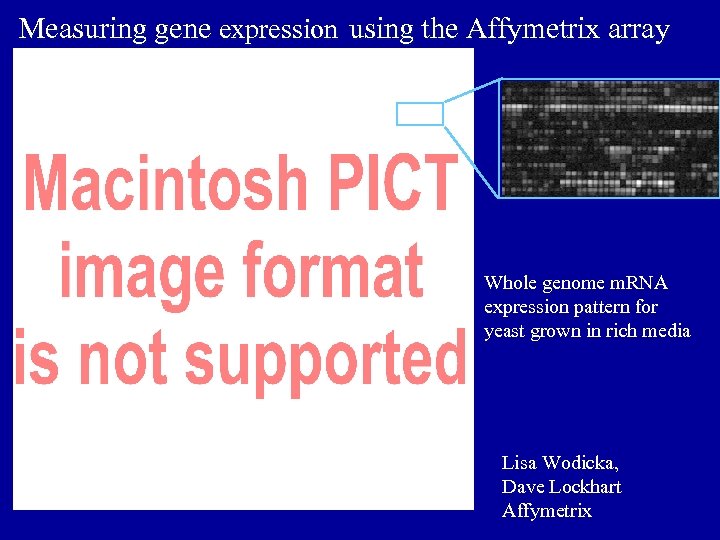 Measuring gene expression using the Affymetrix array Whole genome m. RNA expression pattern for yeast grown in rich media Lisa Wodicka, Dave Lockhart Affymetrix
Measuring gene expression using the Affymetrix array Whole genome m. RNA expression pattern for yeast grown in rich media Lisa Wodicka, Dave Lockhart Affymetrix
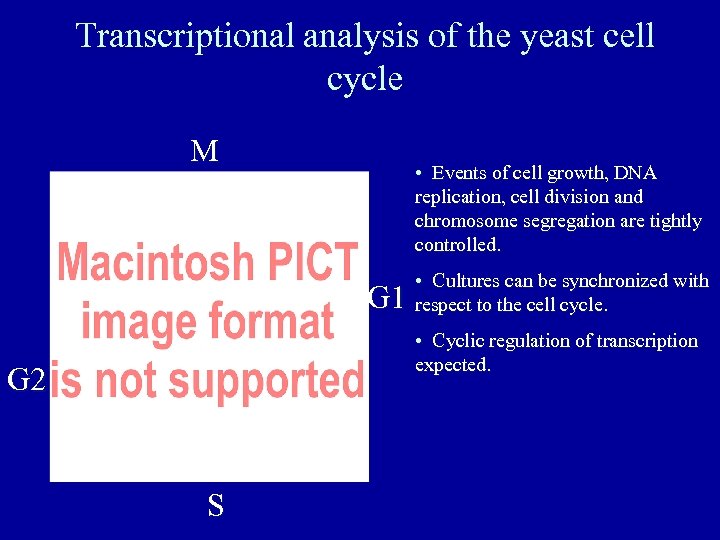 Transcriptional analysis of the yeast cell cycle M • Events of cell growth, DNA replication, cell division and chromosome segregation are tightly controlled. G 1 • Cultures can be synchronized with respect to the cell cycle. • Cyclic regulation of transcription expected. G 2 S
Transcriptional analysis of the yeast cell cycle M • Events of cell growth, DNA replication, cell division and chromosome segregation are tightly controlled. G 1 • Cultures can be synchronized with respect to the cell cycle. • Cyclic regulation of transcription expected. G 2 S
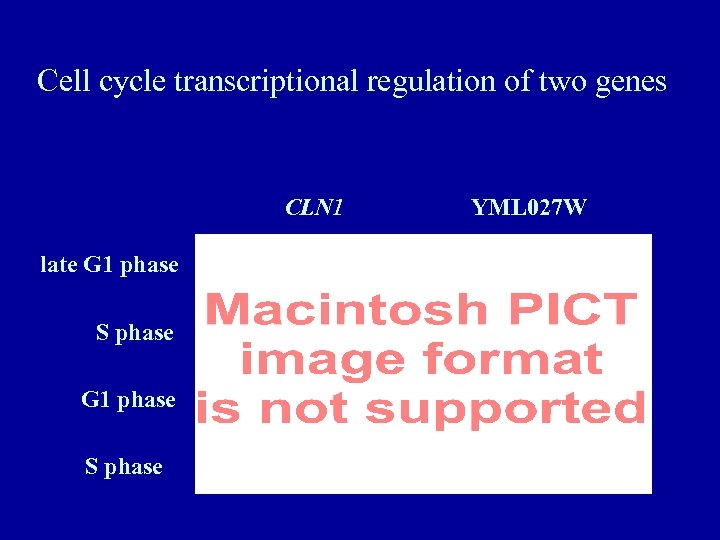 Cell cycle transcriptional regulation of two genes CLN 1 late G 1 phase S phase YML 027 W
Cell cycle transcriptional regulation of two genes CLN 1 late G 1 phase S phase YML 027 W
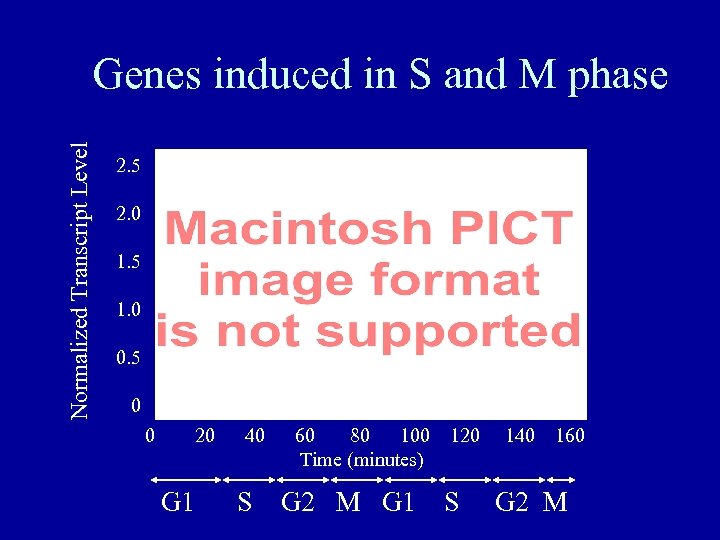 Normalized Transcript Level Genes induced in S and M phase 2. 5 2. 0 1. 5 1. 0 0. 5 0 0 20 G 1 40 60 80 100 120 Time (minutes) S G 2 M G 1 S 140 160 G 2 M
Normalized Transcript Level Genes induced in S and M phase 2. 5 2. 0 1. 5 1. 0 0. 5 0 0 20 G 1 40 60 80 100 120 Time (minutes) S G 2 M G 1 S 140 160 G 2 M
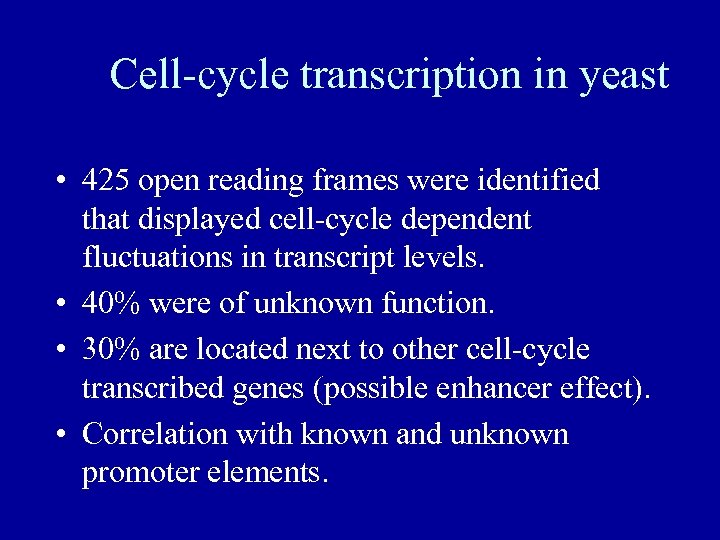 Cell-cycle transcription in yeast • 425 open reading frames were identified that displayed cell-cycle dependent fluctuations in transcript levels. • 40% were of unknown function. • 30% are located next to other cell-cycle transcribed genes (possible enhancer effect). • Correlation with known and unknown promoter elements.
Cell-cycle transcription in yeast • 425 open reading frames were identified that displayed cell-cycle dependent fluctuations in transcript levels. • 40% were of unknown function. • 30% are located next to other cell-cycle transcribed genes (possible enhancer effect). • Correlation with known and unknown promoter elements.
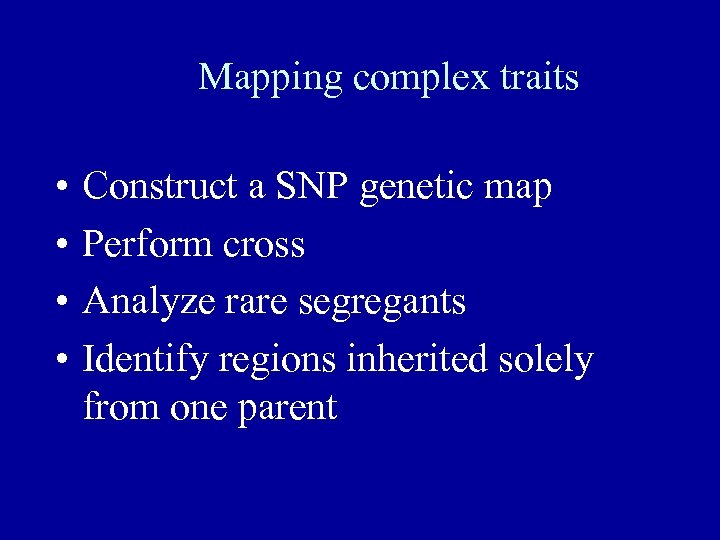 Mapping complex traits • • Construct a SNP genetic map Perform cross Analyze rare segregants Identify regions inherited solely from one parent
Mapping complex traits • • Construct a SNP genetic map Perform cross Analyze rare segregants Identify regions inherited solely from one parent
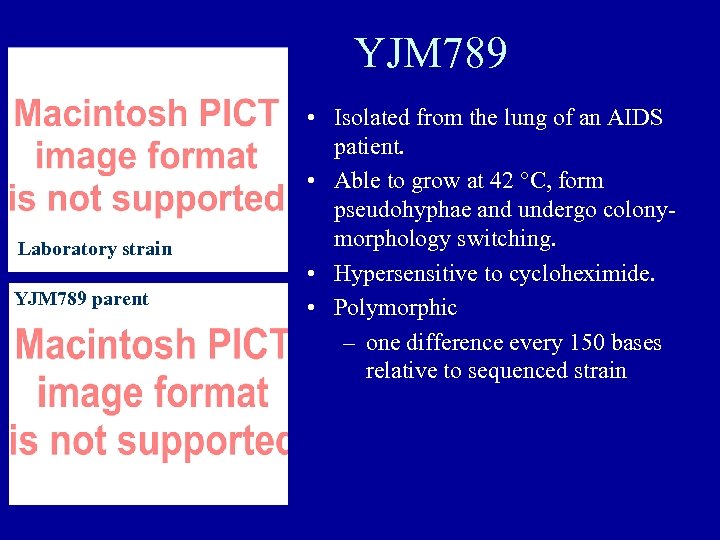 YJM 789 Laboratory strain YJM 789 parent • Isolated from the lung of an AIDS patient. • Able to grow at 42 °C, form pseudohyphae and undergo colonymorphology switching. • Hypersensitive to cycloheximide. • Polymorphic – one difference every 150 bases relative to sequenced strain
YJM 789 Laboratory strain YJM 789 parent • Isolated from the lung of an AIDS patient. • Able to grow at 42 °C, form pseudohyphae and undergo colonymorphology switching. • Hypersensitive to cycloheximide. • Polymorphic – one difference every 150 bases relative to sequenced strain
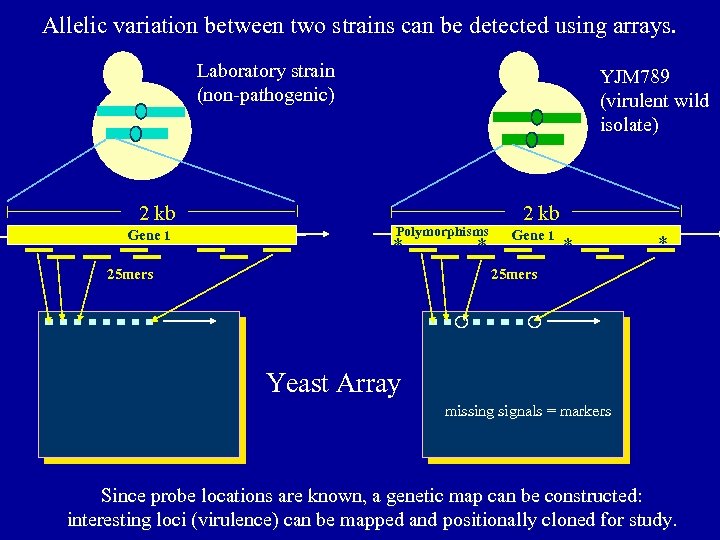 Allelic variation between two strains can be detected using arrays. Laboratory strain (non-pathogenic) 2 kb Gene 1 YJM 789 (virulent wild isolate) Polymorphisms * 25 mers * 2 kb Gene 1 * * 25 mers Mismatch control probe (position 13 of 25) Yeast Array missing signals = markers Since probe locations are known, a genetic map can be constructed: interesting loci (virulence) can be mapped and positionally cloned for study.
Allelic variation between two strains can be detected using arrays. Laboratory strain (non-pathogenic) 2 kb Gene 1 YJM 789 (virulent wild isolate) Polymorphisms * 25 mers * 2 kb Gene 1 * * 25 mers Mismatch control probe (position 13 of 25) Yeast Array missing signals = markers Since probe locations are known, a genetic map can be constructed: interesting loci (virulence) can be mapped and positionally cloned for study.
 Allelic variation in YJM 789 • 3808 markers detected by automated analysis of scanned images. – Largest gap = 56 kb – Average frequency = 3000 bases (1. 0 c. M) • More markers identified in one hybridization than in the past 40 years of yeast genetics.
Allelic variation in YJM 789 • 3808 markers detected by automated analysis of scanned images. – Largest gap = 56 kb – Average frequency = 3000 bases (1. 0 c. M) • More markers identified in one hybridization than in the past 40 years of yeast genetics.
 Verification of markers by tetrad analysis Expect 90 cross-overs per genome. Expect clear recombination breakpoints Expect most markers to segregate 2: 2.
Verification of markers by tetrad analysis Expect 90 cross-overs per genome. Expect clear recombination breakpoints Expect most markers to segregate 2: 2.
 Markers segregate as expected 96 crossovers (90 expected). 96% of markers segregate 2: 2. Clear breakpoints observed. Segregation of markers in one tetrad (one chromosome)
Markers segregate as expected 96 crossovers (90 expected). 96% of markers segregate 2: 2. Clear breakpoints observed. Segregation of markers in one tetrad (one chromosome)
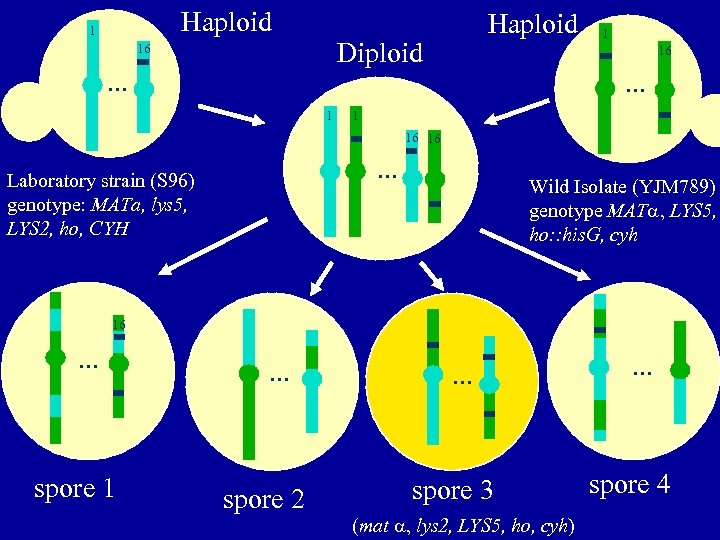 Haploid 1 Haploid Diploid 16 . . . 1 1 16 . . . 1 16 16 . . . Laboratory strain (S 96) genotype: MATa, lys 5, LYS 2, ho, CYH Wild Isolate (YJM 789) genotype MAT LYS 5, ho: : his. G, cyh 16 . . . spore 1 . . . spore 2 . . . spore 3 (mat lys 2, LYS 5, ho, cyh) . . . spore 4
Haploid 1 Haploid Diploid 16 . . . 1 1 16 . . . 1 16 16 . . . Laboratory strain (S 96) genotype: MATa, lys 5, LYS 2, ho, CYH Wild Isolate (YJM 789) genotype MAT LYS 5, ho: : his. G, cyh 16 . . . spore 1 . . . spore 2 . . . spore 3 (mat lys 2, LYS 5, ho, cyh) . . . spore 4
 Inheritance of markers in 10 lys 2 segregants
Inheritance of markers in 10 lys 2 segregants
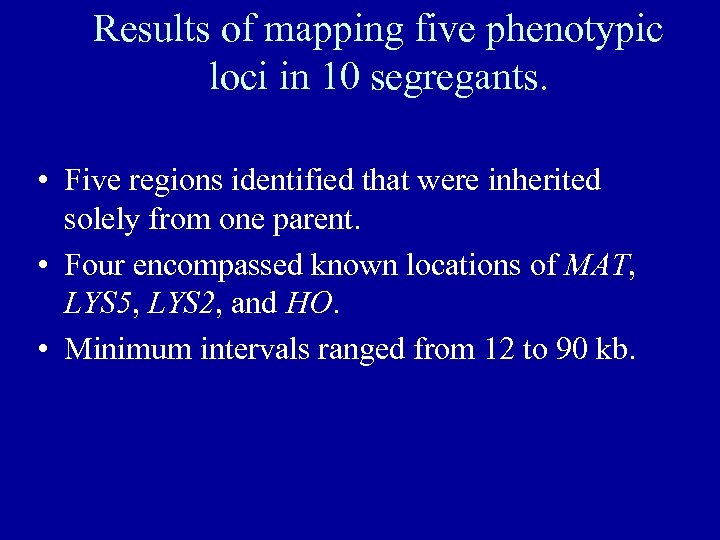 Results of mapping five phenotypic loci in 10 segregants. • Five regions identified that were inherited solely from one parent. • Four encompassed known locations of MAT, LYS 5, LYS 2, and HO. • Minimum intervals ranged from 12 to 90 kb.
Results of mapping five phenotypic loci in 10 segregants. • Five regions identified that were inherited solely from one parent. • Four encompassed known locations of MAT, LYS 5, LYS 2, and HO. • Minimum intervals ranged from 12 to 90 kb.
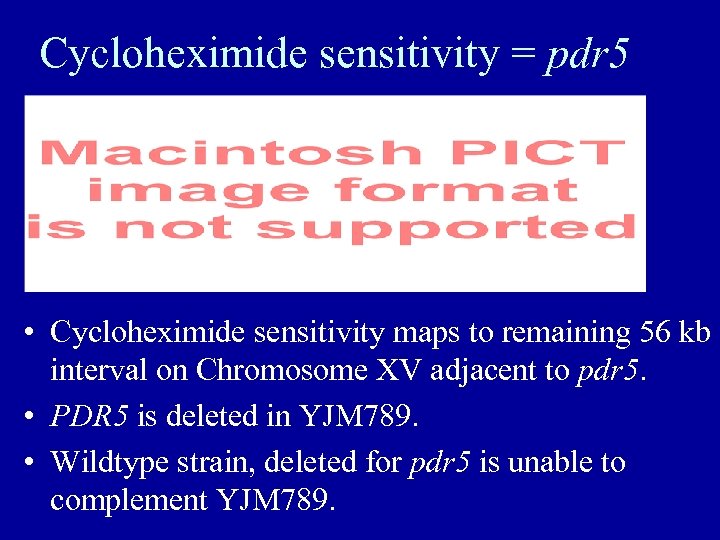 Cycloheximide sensitivity = pdr 5 • Cycloheximide sensitivity maps to remaining 56 kb interval on Chromosome XV adjacent to pdr 5. • PDR 5 is deleted in YJM 789. • Wildtype strain, deleted for pdr 5 is unable to complement YJM 789.
Cycloheximide sensitivity = pdr 5 • Cycloheximide sensitivity maps to remaining 56 kb interval on Chromosome XV adjacent to pdr 5. • PDR 5 is deleted in YJM 789. • Wildtype strain, deleted for pdr 5 is unable to complement YJM 789.
 Mapping Complex Traits: Feasibility Summary • Identified 3808 genetic markers. • Demonstrated that traits can be mapped using these markers. • Next step: Map virulence loci.
Mapping Complex Traits: Feasibility Summary • Identified 3808 genetic markers. • Demonstrated that traits can be mapped using these markers. • Next step: Map virulence loci.
 Virulence in YJM 789 • Virulence is a multigenic trait with 5 loci contributing. – Only 5 of 200 segregants from crosses between YJM 789 and laboratory strain are virulent. • Genes cannot be cloned by complementation. • Hybridization with arrays is an appropriate way to map all contributing loci simultaneously.
Virulence in YJM 789 • Virulence is a multigenic trait with 5 loci contributing. – Only 5 of 200 segregants from crosses between YJM 789 and laboratory strain are virulent. • Genes cannot be cloned by complementation. • Hybridization with arrays is an appropriate way to map all contributing loci simultaneously.
 Assigning Function through Mutational Analysis • Inactivate gene product (delete gene). • Grow mutant strain under different selective or stress conditions. • Identify mutants with growth defects. • Function of gene product may be revealed. – UV sensitivity = DNA repair protein – Adenine auxotrophy = Adenine biosynthesis
Assigning Function through Mutational Analysis • Inactivate gene product (delete gene). • Grow mutant strain under different selective or stress conditions. • Identify mutants with growth defects. • Function of gene product may be revealed. – UV sensitivity = DNA repair protein – Adenine auxotrophy = Adenine biosynthesis
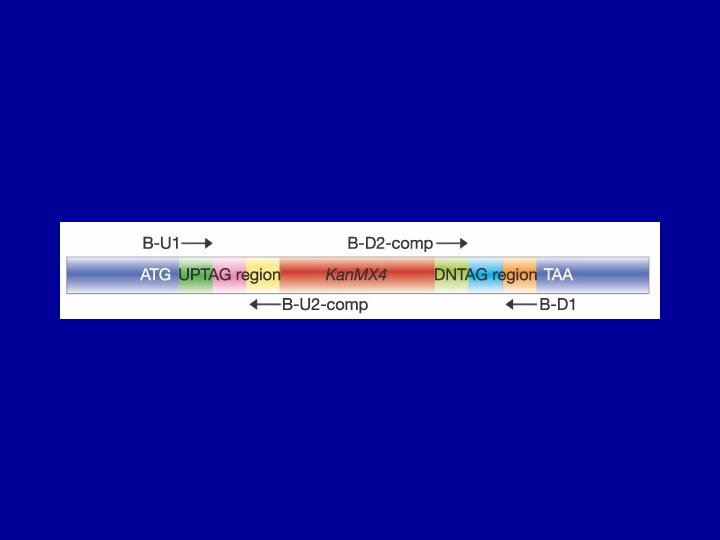
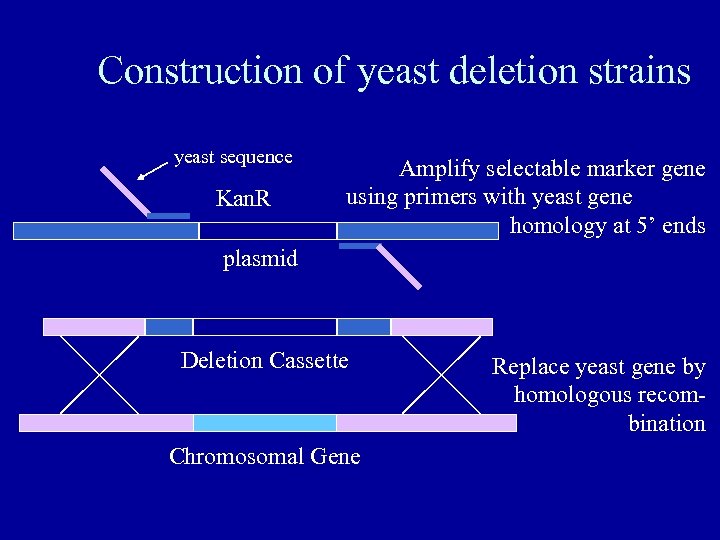 Construction of yeast deletion strains yeast sequence Kan. R Amplify selectable marker gene using primers with yeast gene homology at 5’ ends plasmid Deletion Cassette Chromosomal Gene Replace yeast gene by homologous recombination
Construction of yeast deletion strains yeast sequence Kan. R Amplify selectable marker gene using primers with yeast gene homology at 5’ ends plasmid Deletion Cassette Chromosomal Gene Replace yeast gene by homologous recombination
 International Deletion Consortium Members Mike Snyder, Jasper Rine, Mark Johnston, Jef Boeke, Howard Bussey, Rosetta, Acacia, Peter Philippsen, Hans Hegemann, Francoise Foury, Guido Volckaert, Bruno Andre, Giogio Valle, Jose Revuelta, Steve Kelly, Bart Scherens 24, 000 strains in 3 years
International Deletion Consortium Members Mike Snyder, Jasper Rine, Mark Johnston, Jef Boeke, Howard Bussey, Rosetta, Acacia, Peter Philippsen, Hans Hegemann, Francoise Foury, Guido Volckaert, Bruno Andre, Giogio Valle, Jose Revuelta, Steve Kelly, Bart Scherens 24, 000 strains in 3 years
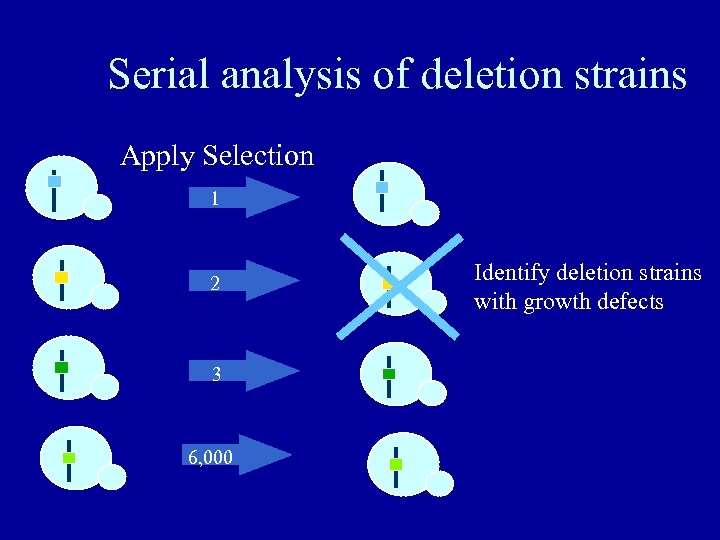 Serial analysis of deletion strains Apply Selection 1 2 3 6, 000 Identify deletion strains with growth defects
Serial analysis of deletion strains Apply Selection 1 2 3 6, 000 Identify deletion strains with growth defects
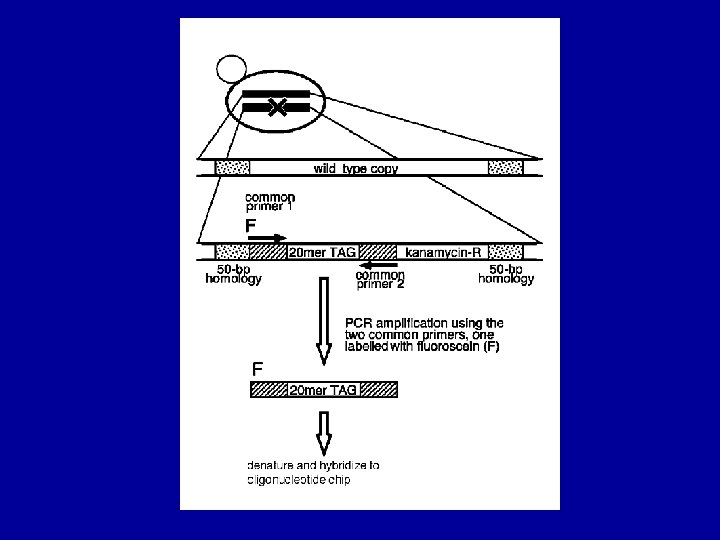
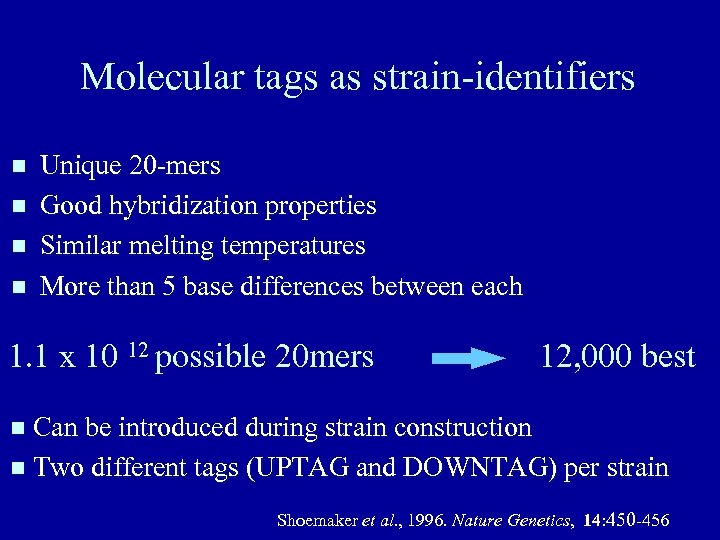 Molecular tags as strain-identifiers Unique 20 -mers Good hybridization properties Similar melting temperatures More than 5 base differences between each 1. 1 x 10 12 possible 20 mers 12, 000 best Can be introduced during strain construction Two different tags (UPTAG and DOWNTAG) per strain Shoemaker et al. , 1996. Nature Genetics, 14: 450 -456
Molecular tags as strain-identifiers Unique 20 -mers Good hybridization properties Similar melting temperatures More than 5 base differences between each 1. 1 x 10 12 possible 20 mers 12, 000 best Can be introduced during strain construction Two different tags (UPTAG and DOWNTAG) per strain Shoemaker et al. , 1996. Nature Genetics, 14: 450 -456
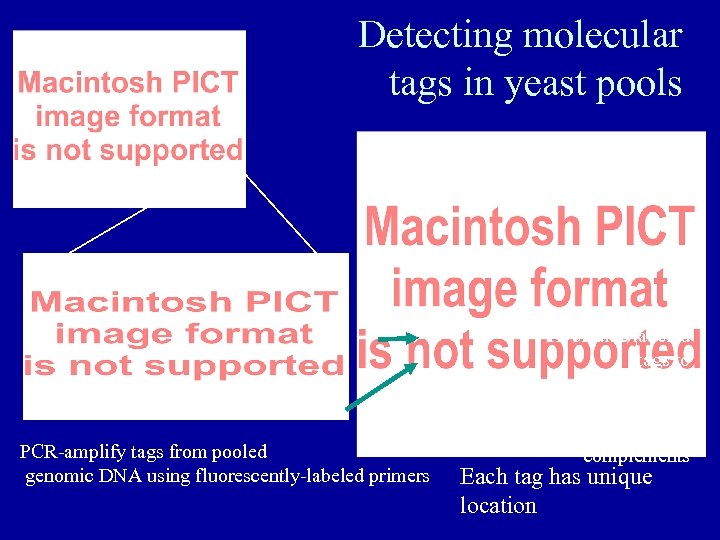 Detecting molecular tags in yeast pools PCR-amplify tags from pooled genomic DNA using fluorescently-labeled primers Hybridize labeled tags to oligonucleotide array containing tag complements Each tag has unique location
Detecting molecular tags in yeast pools PCR-amplify tags from pooled genomic DNA using fluorescently-labeled primers Hybridize labeled tags to oligonucleotide array containing tag complements Each tag has unique location
 Tags can be used to perform negative selections on pools Growth in minimal media identifies all known auxotrophic strains Winzeler et. , 1999 Science 285: 901 -906
Tags can be used to perform negative selections on pools Growth in minimal media identifies all known auxotrophic strains Winzeler et. , 1999 Science 285: 901 -906
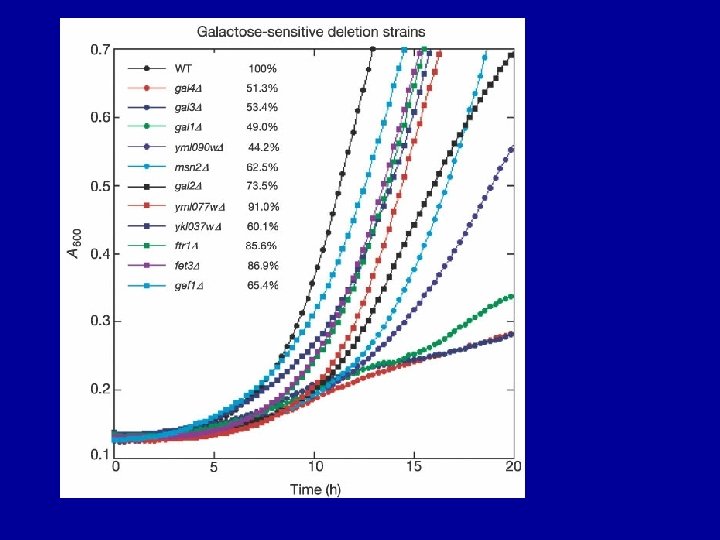
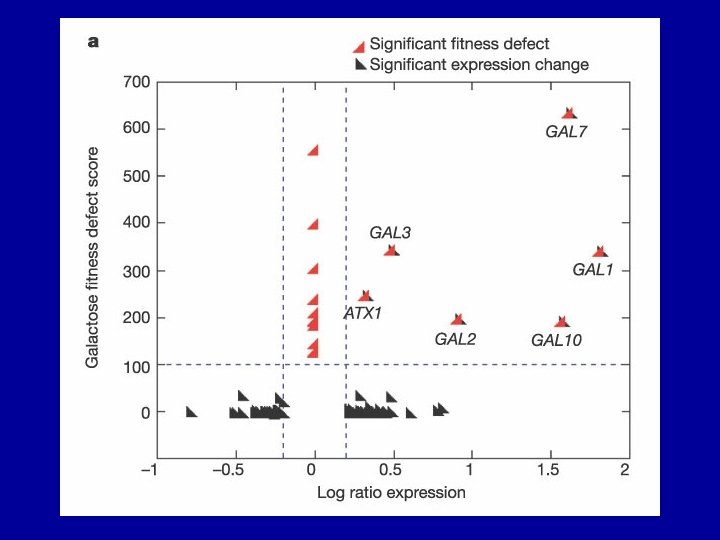
 Genomic profiling of drug sensitivities via “induced haploinsufficiency” Decreased gene dosage from two copies to one copy in heterozygous strains results in increased sensitivity, or drug- induced haploinsufficiency
Genomic profiling of drug sensitivities via “induced haploinsufficiency” Decreased gene dosage from two copies to one copy in heterozygous strains results in increased sensitivity, or drug- induced haploinsufficiency
 Strains that are heterozygous for drug target are haploinsufficient in the presence of drug: Giaever et al. , 1999. Nature Genetics, 21: 278 -283
Strains that are heterozygous for drug target are haploinsufficient in the presence of drug: Giaever et al. , 1999. Nature Genetics, 21: 278 -283
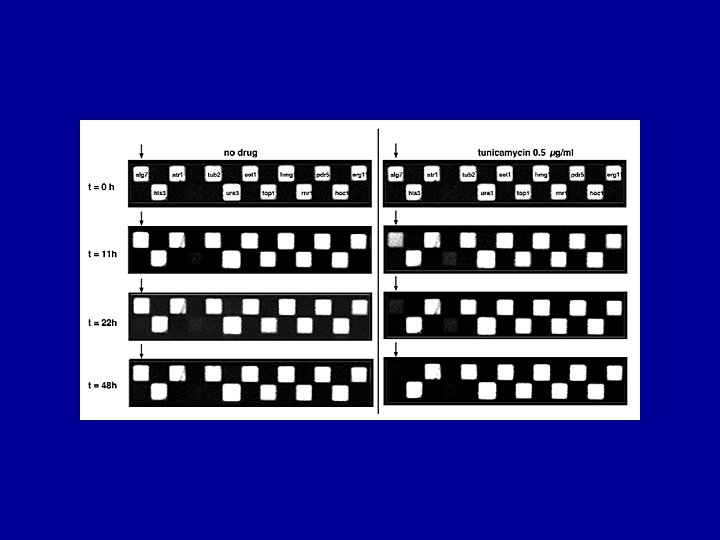
 Tunicamycin sensitivity Analysis of pools of heterozygous (and homozygous) strains reveals primary and secondary drug targets G. Giaever, unpublished results
Tunicamycin sensitivity Analysis of pools of heterozygous (and homozygous) strains reveals primary and secondary drug targets G. Giaever, unpublished results
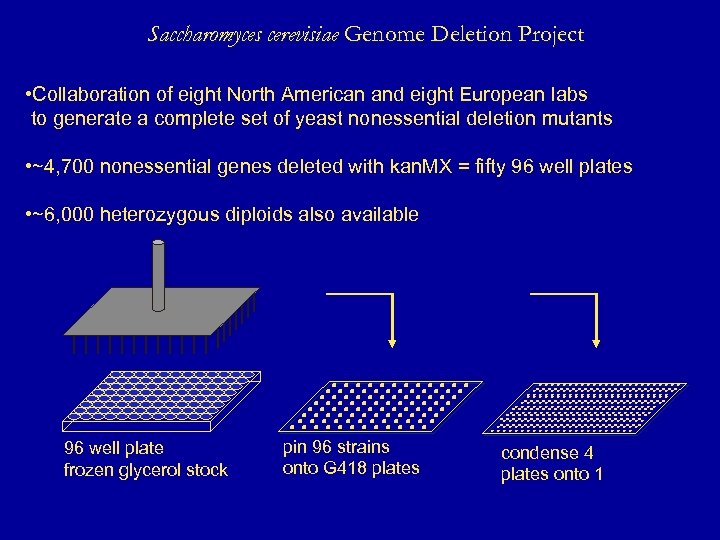 Saccharomyces cerevisiae Genome Deletion Project • Collaboration of eight North American and eight European labs to generate a complete set of yeast nonessential deletion mutants • ~4, 700 nonessential genes deleted with kan. MX = fifty 96 well plates • ~6, 000 heterozygous diploids also available 96 well plate frozen glycerol stock pin 96 strains onto G 418 plates condense 4 plates onto 1
Saccharomyces cerevisiae Genome Deletion Project • Collaboration of eight North American and eight European labs to generate a complete set of yeast nonessential deletion mutants • ~4, 700 nonessential genes deleted with kan. MX = fifty 96 well plates • ~6, 000 heterozygous diploids also available 96 well plate frozen glycerol stock pin 96 strains onto G 418 plates condense 4 plates onto 1
 Examples- global screens Synthetic lethals Synthetic dosage lethals Heterozygous diploids Haploinsufficiency modifiers Increased drug sensitivity- (target ID) Direct phenotype screening
Examples- global screens Synthetic lethals Synthetic dosage lethals Heterozygous diploids Haploinsufficiency modifiers Increased drug sensitivity- (target ID) Direct phenotype screening
 Yeast as a tool to discover drugs and their mechanism of action
Yeast as a tool to discover drugs and their mechanism of action
 Identification of natural compounds that inhibit invasion • Metastasis responsible for 90% of cancer deaths • Metastasis requires invasion of adjacent tissue and blood vessels by tumour cells Lianne Mc. Hardy, Cal Roskelley, Shoki Dedhar, Ali Karsan, David Williams, Raymond Andersen, Michel Roberge
Identification of natural compounds that inhibit invasion • Metastasis responsible for 90% of cancer deaths • Metastasis requires invasion of adjacent tissue and blood vessels by tumour cells Lianne Mc. Hardy, Cal Roskelley, Shoki Dedhar, Ali Karsan, David Williams, Raymond Andersen, Michel Roberge
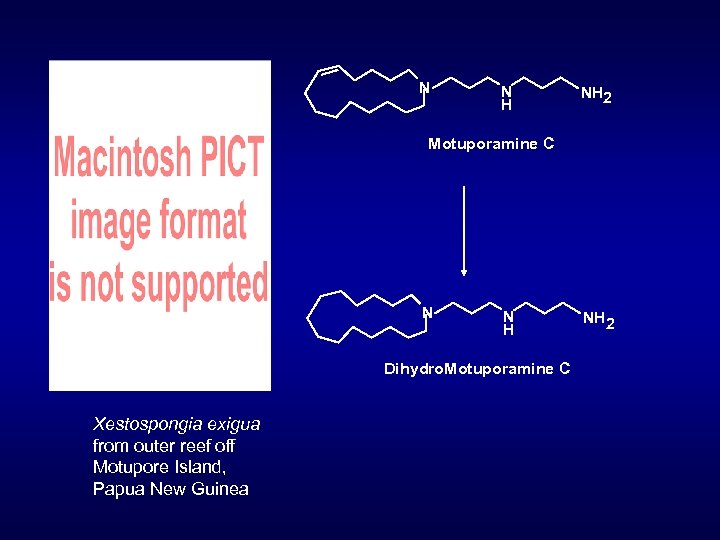 N N H NH 2 Motuporamine C N N H Dihydro. Motuporamine C Xestospongia exigua from outer reef off Motupore Island, Papua New Guinea NH 2
N N H NH 2 Motuporamine C N N H Dihydro. Motuporamine C Xestospongia exigua from outer reef off Motupore Island, Papua New Guinea NH 2
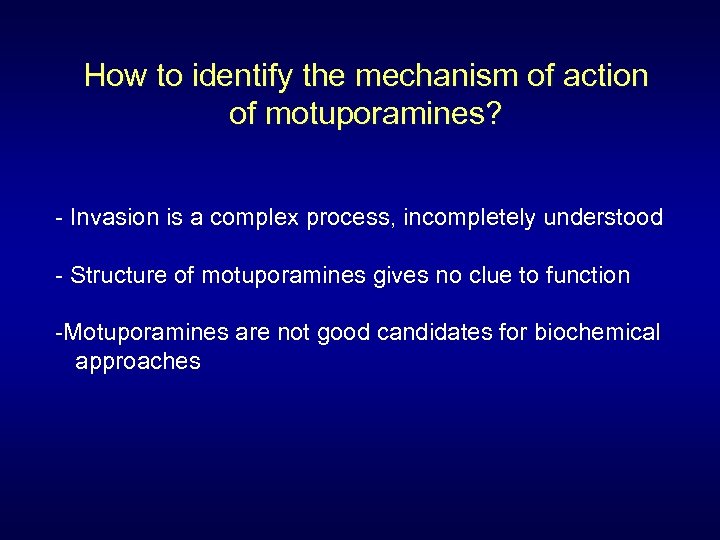 How to identify the mechanism of action of motuporamines? - Invasion is a complex process, incompletely understood - Structure of motuporamines gives no clue to function -Motuporamines are not good candidates for biochemical approaches
How to identify the mechanism of action of motuporamines? - Invasion is a complex process, incompletely understood - Structure of motuporamines gives no clue to function -Motuporamines are not good candidates for biochemical approaches
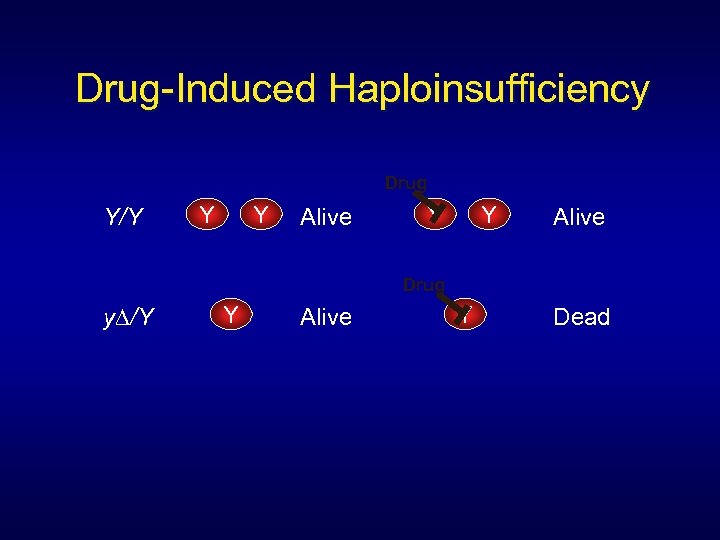 Drug-Induced Haploinsufficiency Drug Y/Y Y Y Alive Drug y∆/Y Y Alive Y Dead
Drug-Induced Haploinsufficiency Drug Y/Y Y Y Alive Drug y∆/Y Y Alive Y Dead
 Drug-induced haploinsufficiency Proof of principle study: Giaever et al. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet 21, 278 -83. (1999) Can these techniques really identify the target or targetted pathways of a drug with an unknown mechanism? Can they predict the target in human cells?
Drug-induced haploinsufficiency Proof of principle study: Giaever et al. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet 21, 278 -83. (1999) Can these techniques really identify the target or targetted pathways of a drug with an unknown mechanism? Can they predict the target in human cells?
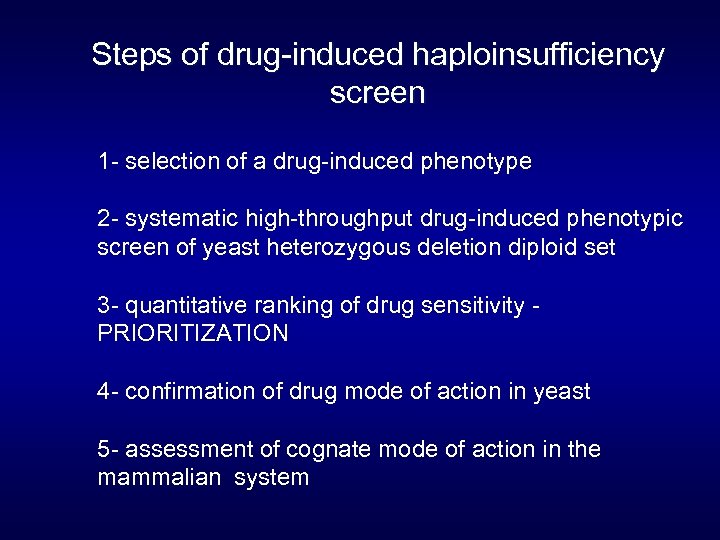 Steps of drug-induced haploinsufficiency screen 1 - selection of a drug-induced phenotype 2 - systematic high-throughput drug-induced phenotypic screen of yeast heterozygous deletion diploid set 3 - quantitative ranking of drug sensitivity PRIORITIZATION 4 - confirmation of drug mode of action in yeast 5 - assessment of cognate mode of action in the mammalian system
Steps of drug-induced haploinsufficiency screen 1 - selection of a drug-induced phenotype 2 - systematic high-throughput drug-induced phenotypic screen of yeast heterozygous deletion diploid set 3 - quantitative ranking of drug sensitivity PRIORITIZATION 4 - confirmation of drug mode of action in yeast 5 - assessment of cognate mode of action in the mammalian system
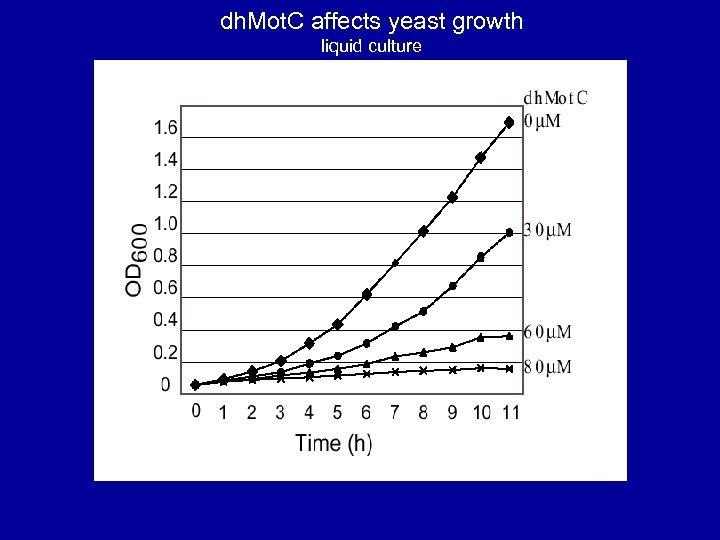 dh. Mot. C affects yeast growth liquid culture
dh. Mot. C affects yeast growth liquid culture
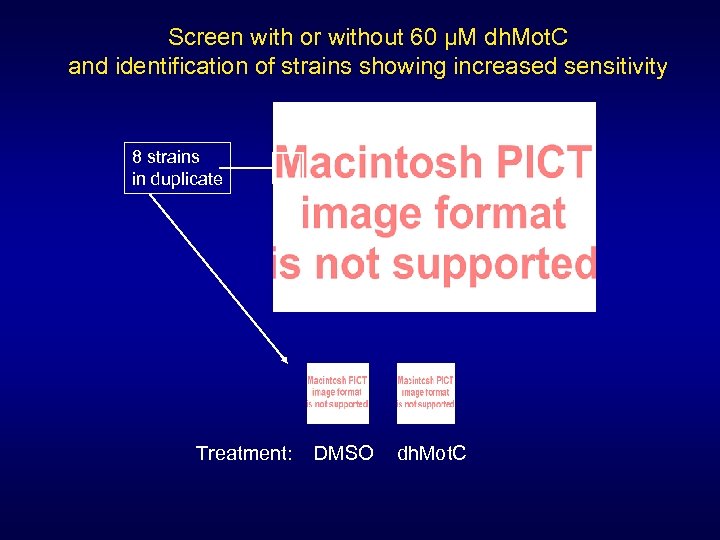 Screen with or without 60 µM dh. Mot. C and identification of strains showing increased sensitivity 8 strains in duplicate Treatment: DMSO dh. Mot. C
Screen with or without 60 µM dh. Mot. C and identification of strains showing increased sensitivity 8 strains in duplicate Treatment: DMSO dh. Mot. C
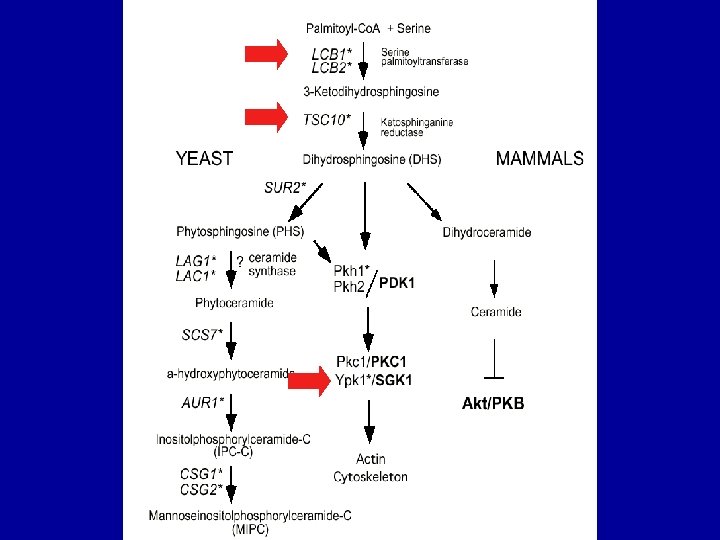
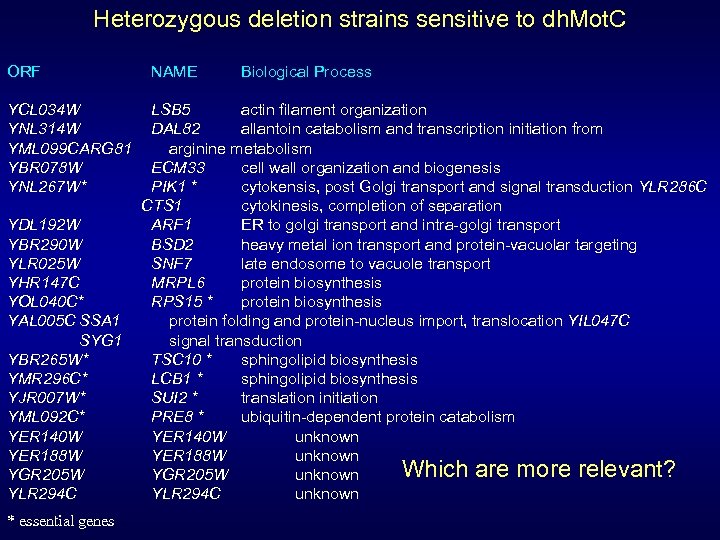 Heterozygous deletion strains sensitive to dh. Mot. C ORF YCL 034 W YNL 314 W YML 099 CARG 81 YBR 078 W YNL 267 W* YDL 192 W YBR 290 W YLR 025 W YHR 147 C YOL 040 C* YAL 005 C SSA 1 SYG 1 YBR 265 W* YMR 296 C* YJR 007 W* YML 092 C* YER 140 W YER 188 W YGR 205 W YLR 294 C * essential genes NAME Biological Process LSB 5 actin filament organization DAL 82 allantoin catabolism and transcription initiation from arginine metabolism ECM 33 cell wall organization and biogenesis PIK 1 * cytokensis, post Golgi transport and signal transduction YLR 286 C CTS 1 cytokinesis, completion of separation ARF 1 ER to golgi transport and intra-golgi transport BSD 2 heavy metal ion transport and protein-vacuolar targeting SNF 7 late endosome to vacuole transport MRPL 6 protein biosynthesis RPS 15 * protein biosynthesis protein folding and protein-nucleus import, translocation YIL 047 C signal transduction TSC 10 * sphingolipid biosynthesis LCB 1 * sphingolipid biosynthesis SUI 2 * translation initiation PRE 8 * ubiquitin-dependent protein catabolism YER 140 W unknown YER 188 W unknown Which are more relevant? YGR 205 W unknown YLR 294 C unknown
Heterozygous deletion strains sensitive to dh. Mot. C ORF YCL 034 W YNL 314 W YML 099 CARG 81 YBR 078 W YNL 267 W* YDL 192 W YBR 290 W YLR 025 W YHR 147 C YOL 040 C* YAL 005 C SSA 1 SYG 1 YBR 265 W* YMR 296 C* YJR 007 W* YML 092 C* YER 140 W YER 188 W YGR 205 W YLR 294 C * essential genes NAME Biological Process LSB 5 actin filament organization DAL 82 allantoin catabolism and transcription initiation from arginine metabolism ECM 33 cell wall organization and biogenesis PIK 1 * cytokensis, post Golgi transport and signal transduction YLR 286 C CTS 1 cytokinesis, completion of separation ARF 1 ER to golgi transport and intra-golgi transport BSD 2 heavy metal ion transport and protein-vacuolar targeting SNF 7 late endosome to vacuole transport MRPL 6 protein biosynthesis RPS 15 * protein biosynthesis protein folding and protein-nucleus import, translocation YIL 047 C signal transduction TSC 10 * sphingolipid biosynthesis LCB 1 * sphingolipid biosynthesis SUI 2 * translation initiation PRE 8 * ubiquitin-dependent protein catabolism YER 140 W unknown YER 188 W unknown Which are more relevant? YGR 205 W unknown YLR 294 C unknown
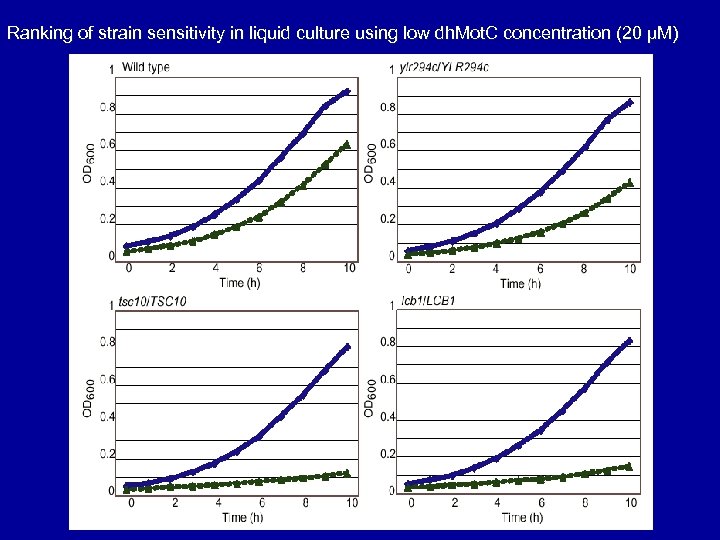 Ranking of strain sensitivity in liquid culture using low dh. Mot. C concentration (20 µM)
Ranking of strain sensitivity in liquid culture using low dh. Mot. C concentration (20 µM)
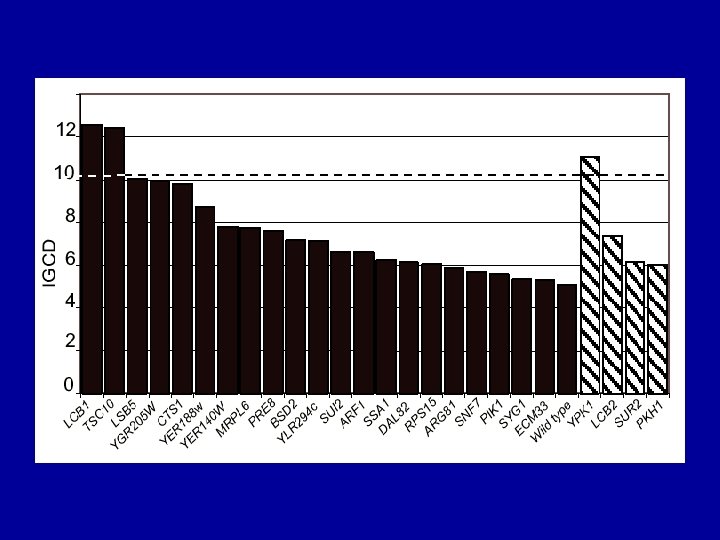
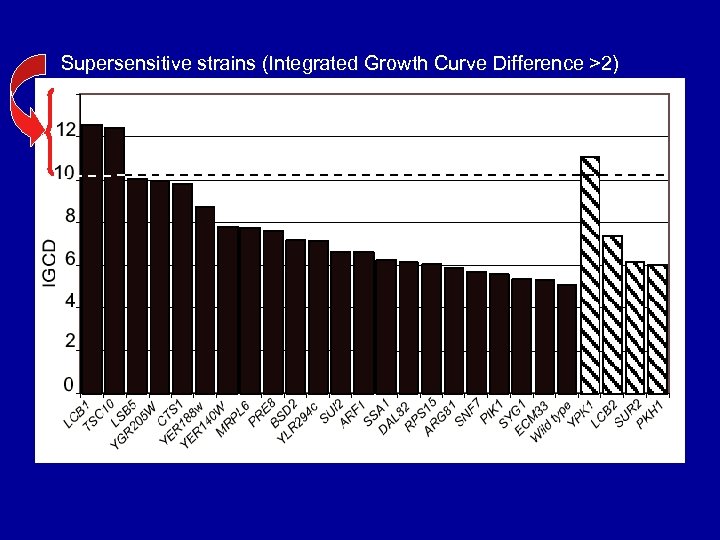 Supersensitive strains (Integrated Growth Curve Difference >2)
Supersensitive strains (Integrated Growth Curve Difference >2)
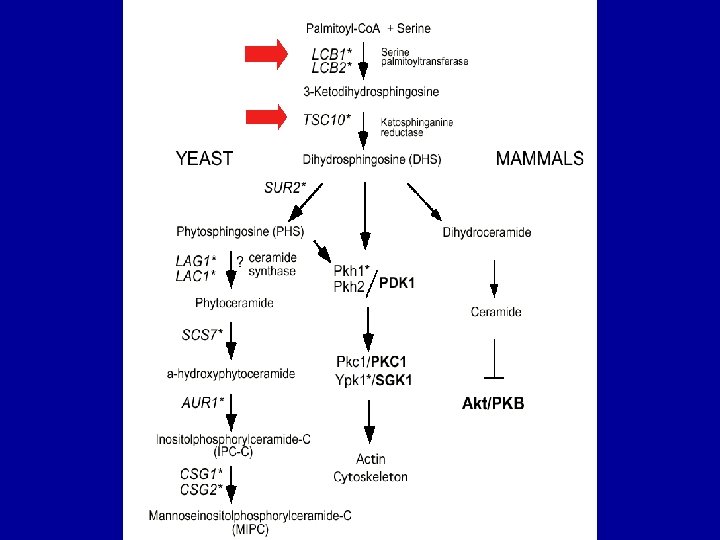
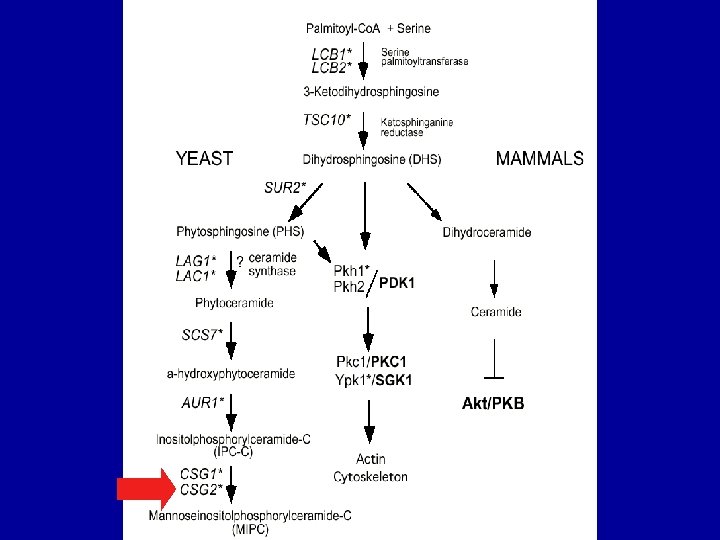
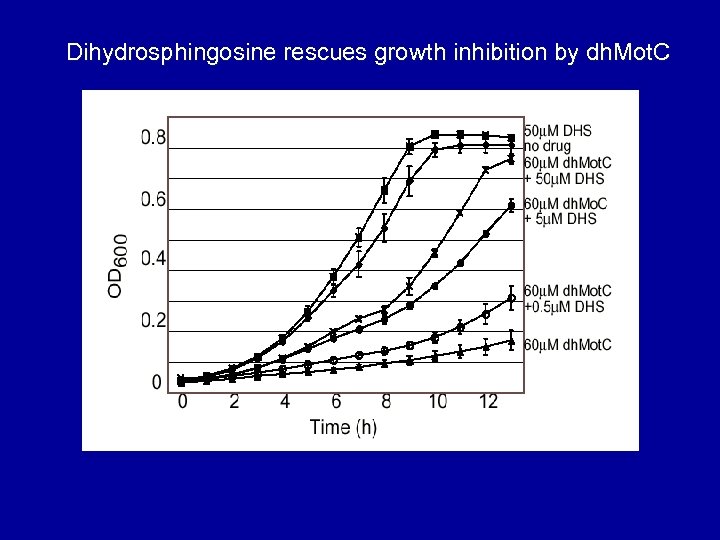 Dihydrosphingosine rescues growth inhibition by dh. Mot. C
Dihydrosphingosine rescues growth inhibition by dh. Mot. C
 Can these techniques really identify the mode of action of a drug? YES Can they predict the target/target pathway in human cells? YES Advantages -systematic, unbiased and genome-wide -adaptable to other phenotypes. -pathway conservation = physiological phenotype -development of chemical probes
Can these techniques really identify the mode of action of a drug? YES Can they predict the target/target pathway in human cells? YES Advantages -systematic, unbiased and genome-wide -adaptable to other phenotypes. -pathway conservation = physiological phenotype -development of chemical probes
 Examples- global screens Synthetic lethals Synthetic dosage lethals Heterozygous diploids Haploinsufficiency modifiers Increased drug sensitivity- (target ID) Direct phenotype screening
Examples- global screens Synthetic lethals Synthetic dosage lethals Heterozygous diploids Haploinsufficiency modifiers Increased drug sensitivity- (target ID) Direct phenotype screening
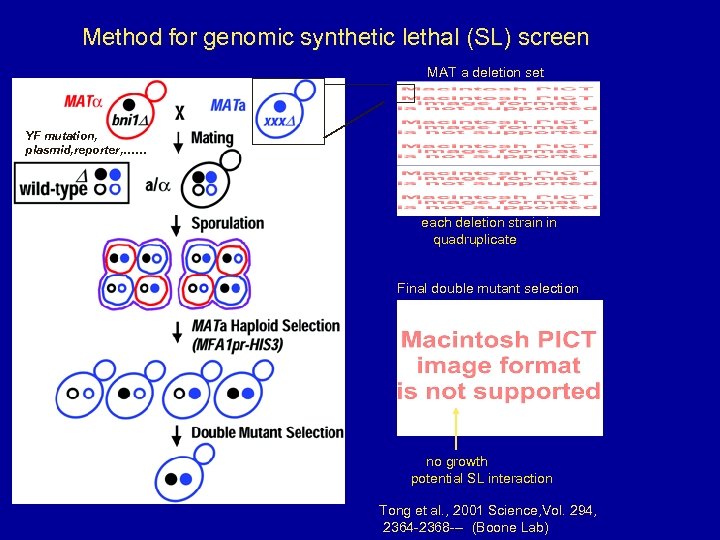 Method for genomic synthetic lethal (SL) screen MAT a deletion set YF mutation, plasmid, reporter, …… each deletion strain in quadruplicate Final double mutant selection no growth potential SL interaction Tong et al. , 2001 Science, Vol. 294, 2364 -2368 --- (Boone Lab)
Method for genomic synthetic lethal (SL) screen MAT a deletion set YF mutation, plasmid, reporter, …… each deletion strain in quadruplicate Final double mutant selection no growth potential SL interaction Tong et al. , 2001 Science, Vol. 294, 2364 -2368 --- (Boone Lab)


 DONE
DONE




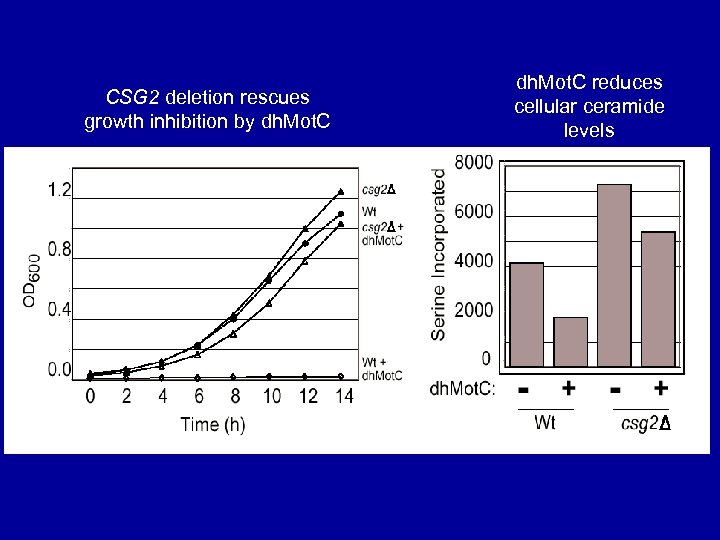 CSG 2 deletion rescues growth inhibition by dh. Mot. C reduces cellular ceramide levels
CSG 2 deletion rescues growth inhibition by dh. Mot. C reduces cellular ceramide levels
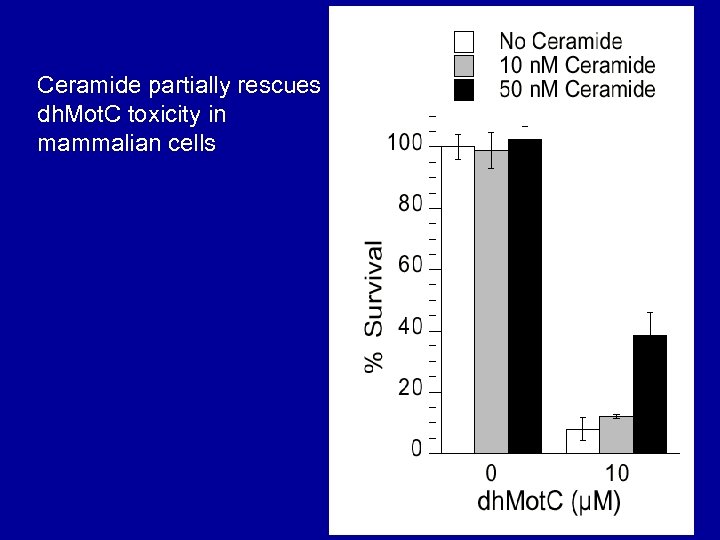 Ceramide partially rescues dh. Mot. C toxicity in mammalian cells
Ceramide partially rescues dh. Mot. C toxicity in mammalian cells


