5c25028e9a2d5eb6bf785056d53f20c8.ppt
- Количество слайдов: 30

Worm. Base : Recent and Future Developments Anthony Rogers* Worm. Base Consortium *Wellcome Trust Sanger Institute California Institute of Technology Cold Spring Harbor Laboratory Washington University at St. Louis

What you told us to do! User survey in Nov/Dec 2005 had 761 respondants http: //www. wormbase. org/announcements/newsletters/pdf/2006 -01. pdf 1) 2) 3) 4) 5) 6) 7) Website navigation and speed Gene structures (see poster P 159) Genetic map (see poster P 157) Phenotypes Literature search Use of other nematode genomes Community forum / wiki

• Web site speed improvements • Extra server hardware • restructured architecture and load balancing • pre-caching popular changes (ie gene pages) • Another European mirror site ! wormbase. sanger. ac. uk

What you told us to do! User survey in Nov/Dec 2005 had 761 respondants http: //www. wormbase. org/announcements/newsletters/pdf/2006 -01. pdf 1) 2) 3) 4) 5) 6) 7) Website navigation and speed Gene structures (see poster P 159) Genetic map (see poster P 157) Phenotypes Literature search Use of other nematode genomes Community forum / wiki

What you told us to do! User survey in Nov/Dec 2005 had 761 respondants http: //www. wormbase. org/announcements/newsletters/pdf/2006 -01. pdf 1) 2) 3) 4) 5) 6) 7) Website navigation and speed Gene structures (see poster P 159) Genetic map (see poster P 157) Phenotypes Literature search Use of other nematode genomes Community forum / wiki
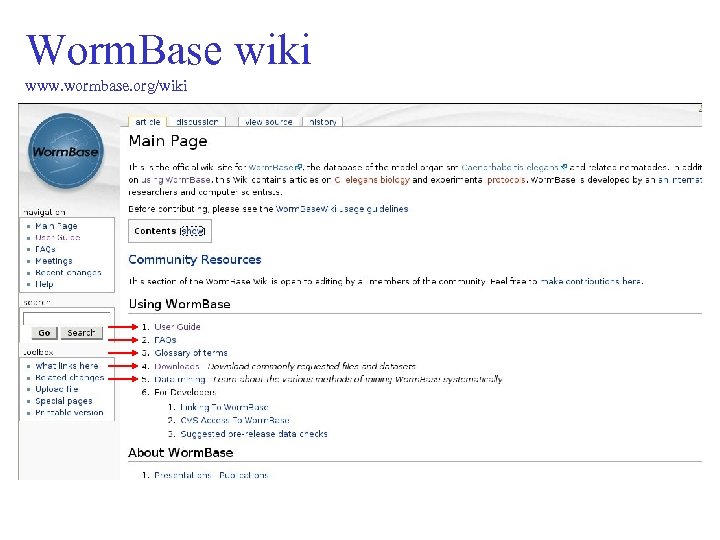
Worm. Base wiki www. wormbase. org/wiki
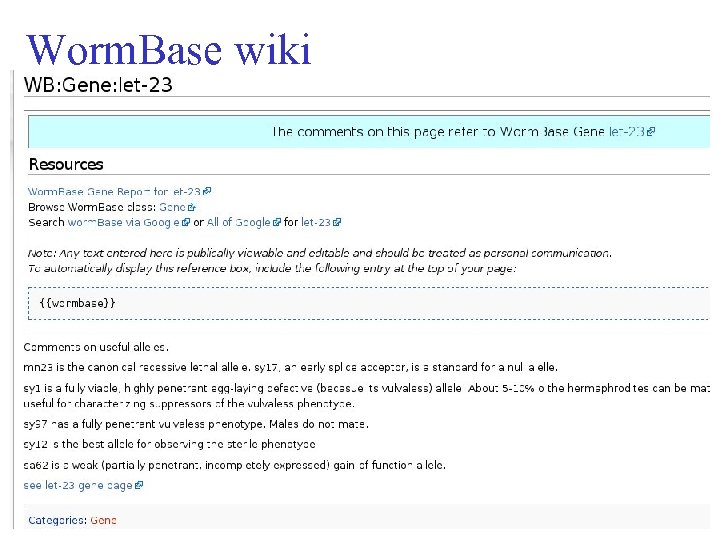
Worm. Base wiki

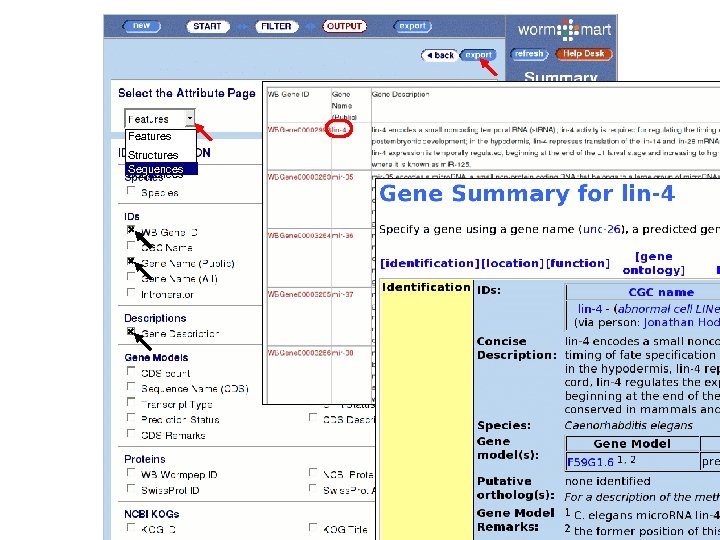
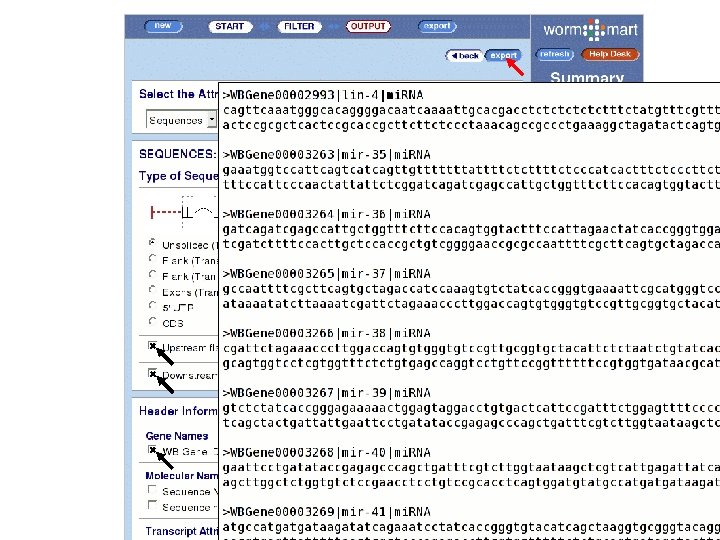
www. wormbase. org/wiki/index. php/Data_mining: Worm. Mart * Example 1 List all synonyms for the following genes; bli-1, egl -43, lag-1. * Example 2 From all genes in C. elegans that have an ortholog in C. briggsae, are located in chromosome III, are sterile in an RNAi screen, and have annotated UTRs, provide a FASTA file containing peptide sequence. * Example 3 Download the set of all RNAi experiments that resulted in an Emb phenotype, and in which the target genes are classified as serine/threonine kinases.

Worm. Mart • Based on the Bio. Mart software • Originally developed at EBI/WTSI for Ensembl, • Various deployments – Worm. Base, Uni. Prot, Gramene. • Worm. Mart • Launched in April 2005, • Replacement for “Batch Genes” and (eventually) “Batch Sequences” pages, • Seven Worm. Base objects are currently described; “Gene”, “GO_term”, “Expression pattern”, “Phenotype”, “RNAi”, “Variation” and “Paper”. • Development is driven largely by user feedback.
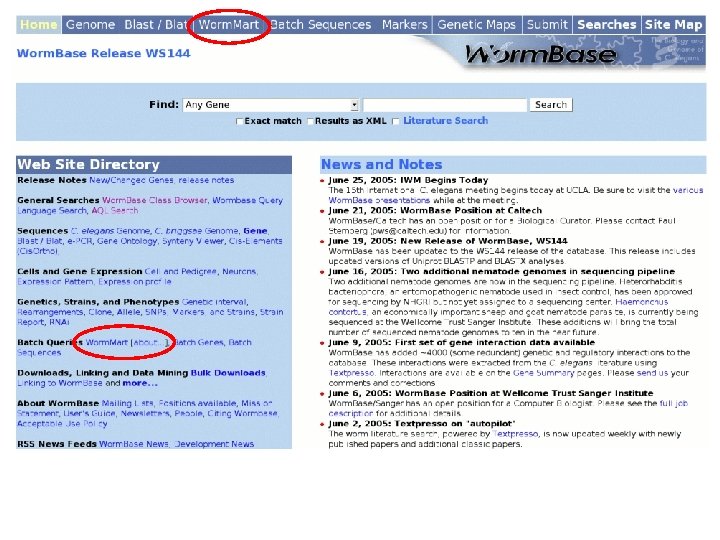
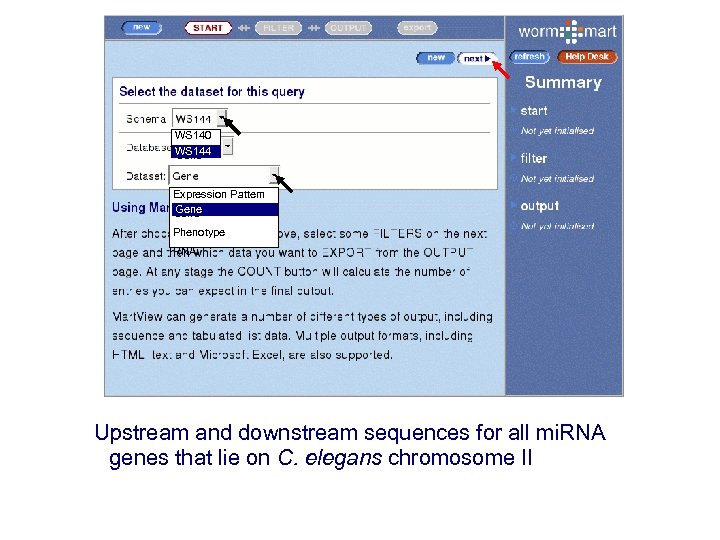
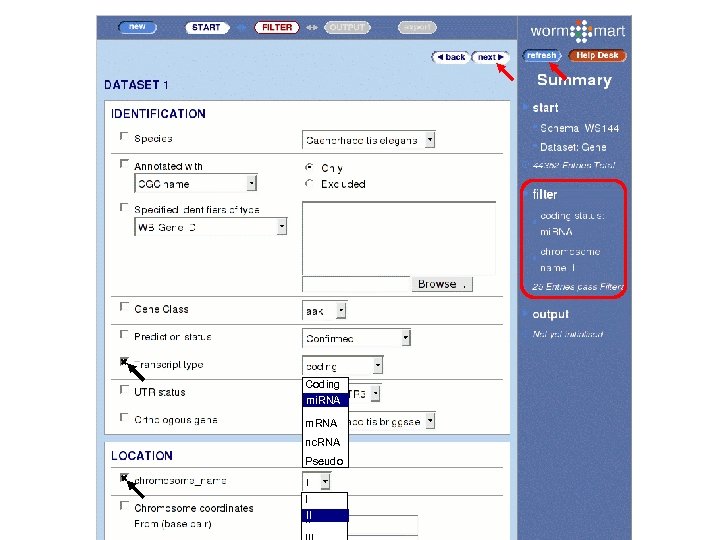
WS 140 WS 144 Gene Expression Pattern Gene Phenotype RNAi Upstream and downstream sequences for all mi. RNA genes that lie on C. elegans chromosome II
Coding mi. RNA mi RNA m. RNA nc. RNA Pseudo I II II
Features Structures Sequences
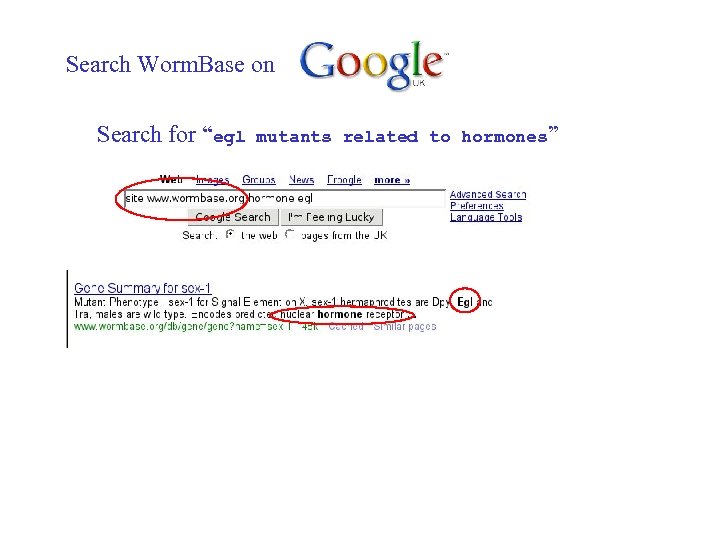
Search Worm. Base on Search for “egl mutants related to hormones”

What you told us to do! User survey in Nov/Dec 2005 had 761 respondants http: //www. wormbase. org/announcements/newsletters/pdf/2006 -01. pdf 1) 2) 3) 4) 5) 6) 7) Website navigation and speed Gene structures (see poster P 159) Genetic map (see poster P 157) Phenotypes Literature search Use of other nematode genomes Community forum / wiki

Comparative genomics • Which species ? • What will we do with them ? • When will this happen ?
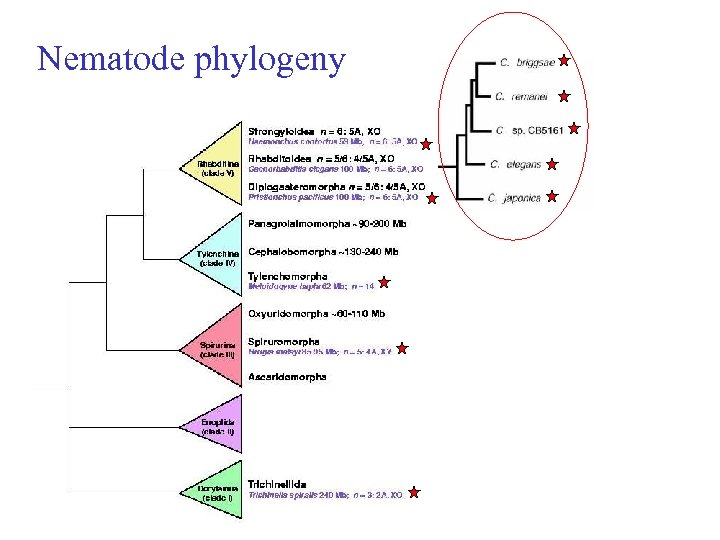
Nematode phylogeny

What we’ll do. . . Pretty much the same as we have with C. briggsae semi-curated gene set based on various predictors Protein set protein annotation ( PFAM, Inter. Pro, tmhmm, signalp ) blastp blastx Whole genome alignment * ortholog assignment *
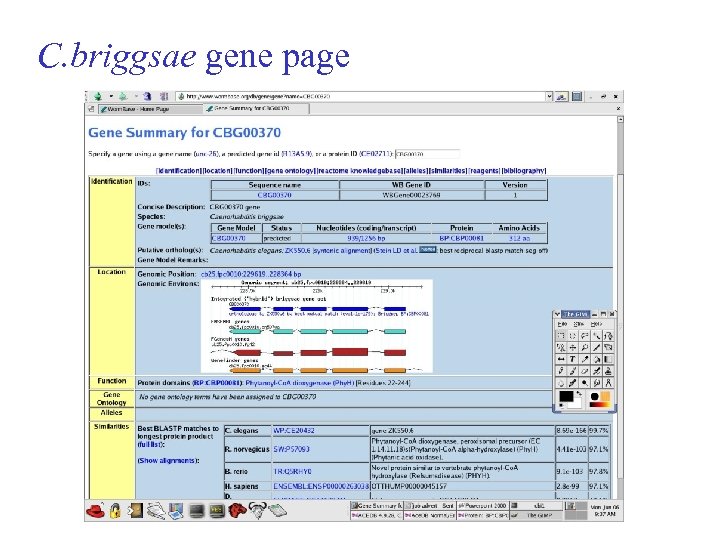
C. briggsae gene page
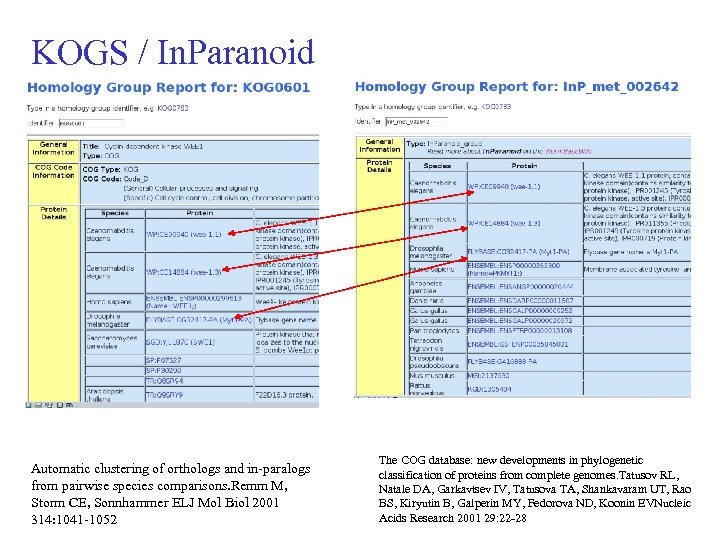
KOGS / In. Paranoid Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. Remm M, Storm CE, Sonnhammer ELJ Mol Biol 2001 314: 1041 -1052 The COG database: new developments in phylogenetic classification of proteins from complete genomes. Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EVNucleic Acids Research 2001 29: 22 -28
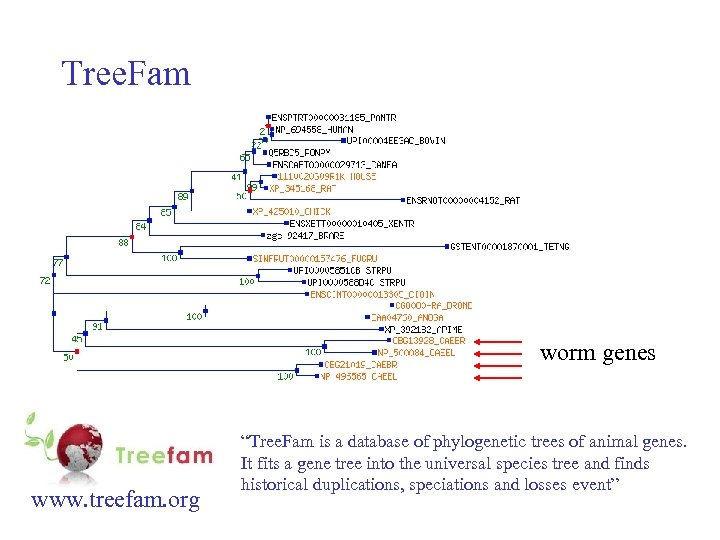
Tree. Fam worm genes www. treefam. org “Tree. Fam is a database of phylogenetic trees of animal genes. It fits a gene tree into the universal species tree and finds historical duplications, speciations and losses event”
Compara “The Ensembl Compara multi-species database stores the results of genome-wide species comparisons calculated for each data release. The database includes Comparative genomics: Whole genome alignments Synteny regions Comparative proteomics: Orthologue predictions Paralogue predictions Protein family clusters” http: //www. ensembl. org/info/software/compara/index. html

What you told us to do! User survey in Nov/Dec 2005 had 761 respondants http: //www. wormbase. org/announcements/newsletters/pdf/2006 -01. pdf 1) 2) 3) 4) 5) 6) 7) Website navigation and speed Gene structures (see poster P 159) Genetic map (see poster P 157) Phenotypes Literature search Use of other nematode genomes Community forum / wiki

Phenotype ontology • Developing a controlled vocabulary for the description of phenotpyes • Will allow high level and more detailed descriptions to be contained in a hierarchial, browsable structure. • Fine grained enough to distinguish between specific experimental definitions of a phenotype if required.
Vancouver Fosmids Sequence report page • lists Transcripts and Microarray assays falling within the span of each fosmid. • interpolated genetic map position • DNA sequence • Options to expand lists of EST, waba and blast alignments, Repeats and RNAi expts. • Link to order from http: //www. geneservice. co. uk/products/clones/Celegans_Fos. jsp

Worm. Book “Worm. Book is a comprehensive, open-access collection of original, peer-reviewed chapters covering topics related to the biology of Caenorhabditis elegans (C. elegans). Worm. Book also includes Worm. Methods, an up-to-date collection of methods and protocols for C. elegans researchers. ” Currently hold 107 chapters. www. wormbook. org wormbook. sanger. ac. uk

Useful files you may not know about best_blastp_hits. WS 157. gz -- Best blast. P hit for each worm protein CE 00081, WP: CE 24153, 4. 8 e-121, ENSEMBL: ENSP 00000320546, 4 e-07, BP: CBP 23671, 4 e-117, FLYBASE: CG 7971 -PD, 2. 3 e-06 *oligo_mapping. gz for affy, agilent and gsc chips ( 3 files ) Oligo_set WBGene. ID Gene_sequence_name Gene_type Microarray_type cea 2. 3. 00017 WBGene 00044022 AC 3. 9 CDS GSC at Wash. U cdna 2 orf. WS 157. gz c. DNA CDS yk 1288 c 01. 3, H 22 K 11. 1 confirmed_genes. WS 157. gz - FASTA fomat file of CDSs with full transcript evidence gene. IDs. WS 157. gz Gene_id WBGene 00000012, CGC name Seq name abf-1, C 50 F 2. 9 pcr_product 2 gene. WS 157. gz pcr_product Gene_id (cgc_name) sjj_C 55 H 1. 2 WBGene 00001672(gpa-10), Seq name C 55 H 1. 2 intergenic_sequences. dna. gz >Gene_id_Gene_id Chromosome Start coord length >WBGene 00022276_WBGene 00022278 CHROMOSOME_I 16832, len: 687 atgttggcaggttttttcagtagtttttgagtgaaaatagaggtaaaaagacagaaaatc aataaaaaatgaaaactatgaaaaatggttgaaaatcgagcaaaaatcgttcaaa

Why isn’t data from paper X in Worm. Base ? • List of emails and forms where data can be submitted. • User submitted data is PRIORITISED over normal curation pipelines • For large or novel data sets contact us asap - before publication - confidentiality agreed wormbase-help@wormbase. org or www. wormbase. org/db/misc/feedback

California Institute of Technology Igor Antoshechkin Carol Bastiani Juancarlos Chan Wen Chen Ranjana Kishore Raymond Lee Hans-Michael Mueller Cecilia Nakamura Andrei Petcherski Gary Schindelman Erich Schwarz Paul Sternberg Kimberly Van Auken Daniel Wang Washington University at St. Louis Tamberlyn Bieri Darin Blasiar Phil Ozersky John Spieth Wellcome Trust Sanger Institute Paul Davis Richard Durbin Michael Han Anthony Rogers Mary Ann Tuli Gary Williams Cold Spring Harbor Laboratory Payan Canaran Jack Chen Tristan Fiedler Todd Harris Sheldon Mc. Kay Will Spooner Lincoln Stein
5c25028e9a2d5eb6bf785056d53f20c8.ppt