a4295df0be435331abce0f9a12414f45.ppt
- Количество слайдов: 29
 What’s New in CCP 4 6. 0? Martyn Winn CCP 4 CCLRC Daresbury Laboratory Cheshire CCP 4 helpdesk: ccp 4@dl. ac. uk
What’s New in CCP 4 6. 0? Martyn Winn CCP 4 CCLRC Daresbury Laboratory Cheshire CCP 4 helpdesk: ccp 4@dl. ac. uk
 Background CCP 4 helpdesk: ccp 4@dl. ac. uk
Background CCP 4 helpdesk: ccp 4@dl. ac. uk
 Collaborative Computational Project Number 4 CCP 4 software suite: • Scope: covers data processing through to refinement and validation • Modular: lots of individual programs sharing data via common file formats • Keywords control program function and provide additional data • Redundancy in choice of programs • Also has a graphical interface CCP 4 i Version 6. 0 was released in February 2006 Version 6. 0. 1 was released in June 2006 CCP 4 website: http: //www. ccp 4. ac. uk/ Old workshops and course: http: //www. ccp 4. ac. uk/ccp 4 course. php Bulletin Board http: //www. ccp 4. ac. uk/ccp 4 bb. php Newsletters: http: //www. ccp 4. ac. uk/newsletters. php CCP 4 helpdesk: ccp 4@dl. ac. uk
Collaborative Computational Project Number 4 CCP 4 software suite: • Scope: covers data processing through to refinement and validation • Modular: lots of individual programs sharing data via common file formats • Keywords control program function and provide additional data • Redundancy in choice of programs • Also has a graphical interface CCP 4 i Version 6. 0 was released in February 2006 Version 6. 0. 1 was released in June 2006 CCP 4 website: http: //www. ccp 4. ac. uk/ Old workshops and course: http: //www. ccp 4. ac. uk/ccp 4 course. php Bulletin Board http: //www. ccp 4. ac. uk/ccp 4 bb. php Newsletters: http: //www. ccp 4. ac. uk/newsletters. php CCP 4 helpdesk: ccp 4@dl. ac. uk
 Scope of the CCP 4 Suite • ~200 (? ) programs covering all aspects of PX structure determination • Relatively small subset are significant for most users, e. g. : • Data processing and reduction (mosflm, scala, truncate. . . ) • Experimental phasing (mlphare, acorn, rantan, oasis, bp 3. . . ) • Molecular replacement (amore, molrep, phaser) • Density modification (dm, solomon, pirate) • Refinement (refmac 5, sketcher) • Graphics and building (ccp 4 mg, coot) • Validation and analysis (sfcheck, procheck …) • Utility and conversion programs (e. g. fft, cad, revise …) CCP 4 helpdesk: ccp 4@dl. ac. uk
Scope of the CCP 4 Suite • ~200 (? ) programs covering all aspects of PX structure determination • Relatively small subset are significant for most users, e. g. : • Data processing and reduction (mosflm, scala, truncate. . . ) • Experimental phasing (mlphare, acorn, rantan, oasis, bp 3. . . ) • Molecular replacement (amore, molrep, phaser) • Density modification (dm, solomon, pirate) • Refinement (refmac 5, sketcher) • Graphics and building (ccp 4 mg, coot) • Validation and analysis (sfcheck, procheck …) • Utility and conversion programs (e. g. fft, cad, revise …) CCP 4 helpdesk: ccp 4@dl. ac. uk
 Graphical User Interface CCP 4 i • Sits on top of programs • Hides details of individual programs • User not locked in • Allows mix-and-match approach • Philosophy: • “Task-driven” rather than “program-driven” • Interfaces to tasks use “natural language” approach • e. g. • Key features: • Easy-to-use interfaces to major programs and utilities • Integrated tools for file viewing and basic project management • Customisable • Integrated help system • Some non-CCP 4 programs also interfaced (e. g. ARP/w. ARP, SHELX) CCP 4 helpdesk: ccp 4@dl. ac. uk
Graphical User Interface CCP 4 i • Sits on top of programs • Hides details of individual programs • User not locked in • Allows mix-and-match approach • Philosophy: • “Task-driven” rather than “program-driven” • Interfaces to tasks use “natural language” approach • e. g. • Key features: • Easy-to-use interfaces to major programs and utilities • Integrated tools for file viewing and basic project management • Customisable • Integrated help system • Some non-CCP 4 programs also interfaced (e. g. ARP/w. ARP, SHELX) CCP 4 helpdesk: ccp 4@dl. ac. uk
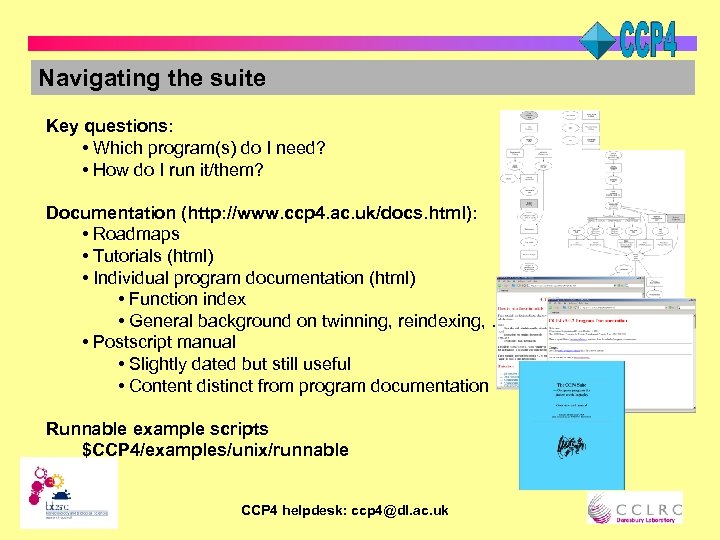 Navigating the suite Key questions: • Which program(s) do I need? • How do I run it/them? Documentation (http: //www. ccp 4. ac. uk/docs. html): • Roadmaps • Tutorials (html) • Individual program documentation (html) • Function index • General background on twinning, reindexing, … • Postscript manual • Slightly dated but still useful • Content distinct from program documentation Runnable example scripts $CCP 4/examples/unix/runnable CCP 4 helpdesk: ccp 4@dl. ac. uk
Navigating the suite Key questions: • Which program(s) do I need? • How do I run it/them? Documentation (http: //www. ccp 4. ac. uk/docs. html): • Roadmaps • Tutorials (html) • Individual program documentation (html) • Function index • General background on twinning, reindexing, … • Postscript manual • Slightly dated but still useful • Content distinct from program documentation Runnable example scripts $CCP 4/examples/unix/runnable CCP 4 helpdesk: ccp 4@dl. ac. uk
 Automated download and install for 6. 0 Downloads divided into a number of packages: • Core CCP 4 (equivalent to v 5. 0) • Phaser and CCTBX • CCP 4 mg • Coot • CHOOCH • plus dependencies (“tools”): Tcl/Tk/BLT, Python Installation: • For Macintosh and Windows: use the self-extracting packages • On Windows: • remove any previous installation first • admin privileges are required to install • For Linux, Unix variants • use install. sh script in download archive to install automatically
Automated download and install for 6. 0 Downloads divided into a number of packages: • Core CCP 4 (equivalent to v 5. 0) • Phaser and CCTBX • CCP 4 mg • Coot • CHOOCH • plus dependencies (“tools”): Tcl/Tk/BLT, Python Installation: • For Macintosh and Windows: use the self-extracting packages • On Windows: • remove any previous installation first • admin privileges are required to install • For Linux, Unix variants • use install. sh script in download archive to install automatically
 CCP 4 6. 0 CCP 4 helpdesk: ccp 4@dl. ac. uk
CCP 4 6. 0 CCP 4 helpdesk: ccp 4@dl. ac. uk
 Updated programs in core CCP 4 6. 0 New versions of major programs: • REFMAC 5. 2. 0019 • MOLREP 9. 0. 09 • SFCHECK 7. 0. 18 • MOSFLM 6. 2. 5 Updates to minor programs: • PDBCUR (PDB editing tool): Summarise model content, choose most likely conformation, delete hydrogens/atoms based on occupancy threshold … • MATTHEWS_COEFF: Outputs Kantardjieff & Rupp resolution-based probabilities for number of molecules in ASU CCP 4 helpdesk: ccp 4@dl. ac. uk
Updated programs in core CCP 4 6. 0 New versions of major programs: • REFMAC 5. 2. 0019 • MOLREP 9. 0. 09 • SFCHECK 7. 0. 18 • MOSFLM 6. 2. 5 Updates to minor programs: • PDBCUR (PDB editing tool): Summarise model content, choose most likely conformation, delete hydrogens/atoms based on occupancy threshold … • MATTHEWS_COEFF: Outputs Kantardjieff & Rupp resolution-based probabilities for number of molecules in ASU CCP 4 helpdesk: ccp 4@dl. ac. uk
 New programs in core CCP 4 Superpose (Eugene Krissinel, EBI) • secondary-structure based alignment program • aligns first on secondary structure then on Cα atom positions • outputs the transformation matrix plus per-residue listing of the quality of the match & identification of the secondary structure BP 3 (Raj Pannu, Leiden) • heavy-atom refinement and phasing program • uses multivariate likelihood functions to refine and phase heavy and/or anomalously scattering substructure (i. e. S/MIR(AS) and S/MAD) Pdb_merge • “Jiffy” program to combine the content from two PDB files CCP 4 helpdesk: ccp 4@dl. ac. uk
New programs in core CCP 4 Superpose (Eugene Krissinel, EBI) • secondary-structure based alignment program • aligns first on secondary structure then on Cα atom positions • outputs the transformation matrix plus per-residue listing of the quality of the match & identification of the secondary structure BP 3 (Raj Pannu, Leiden) • heavy-atom refinement and phasing program • uses multivariate likelihood functions to refine and phase heavy and/or anomalously scattering substructure (i. e. S/MIR(AS) and S/MAD) Pdb_merge • “Jiffy” program to combine the content from two PDB files CCP 4 helpdesk: ccp 4@dl. ac. uk
 New core programs: pirate Kevin Cowtan, York Clipper • a set of crystallographic software libraries developed by Kevin Cowtan • see http: //www. ysbl. york. ac. uk/~cowtan Pirate • statistical phase improvement (replacement for dm) • Nb: executables are actually called cpirate and cmakereference Other Clipper utilities in 6. 0 • see e. g. the Clipper Utilities module of CCP 4 i CCP 4 helpdesk: ccp 4@dl. ac. uk
New core programs: pirate Kevin Cowtan, York Clipper • a set of crystallographic software libraries developed by Kevin Cowtan • see http: //www. ysbl. york. ac. uk/~cowtan Pirate • statistical phase improvement (replacement for dm) • Nb: executables are actually called cpirate and cmakereference Other Clipper utilities in 6. 0 • see e. g. the Clipper Utilities module of CCP 4 i CCP 4 helpdesk: ccp 4@dl. ac. uk
 New core programs: pirate • General categorisation of protein/solvent (order/disorder vs dense/sparse) • Uses phases from known reference structure with features similar to unknown structure (use makereference program for this) • Doesn’t require knowledge of solvent content • Pirate NCS. Novel feature: each molecule gets its own mask based on agreement with NCS. • Pirate in 6. 0. Slower than DM. Dependent on quality of HL coeffs, because they describe noise used to generate noisy reference structure. • For large reference structures, not so sensitive to choice. Might want something else for protein-DNA. Otherwise 1 ajr. • New version of pirate in August CCP 4 helpdesk: ccp 4@dl. ac. uk
New core programs: pirate • General categorisation of protein/solvent (order/disorder vs dense/sparse) • Uses phases from known reference structure with features similar to unknown structure (use makereference program for this) • Doesn’t require knowledge of solvent content • Pirate NCS. Novel feature: each molecule gets its own mask based on agreement with NCS. • Pirate in 6. 0. Slower than DM. Dependent on quality of HL coeffs, because they describe noise used to generate noisy reference structure. • For large reference structures, not so sensitive to choice. Might want something else for protein-DNA. Otherwise 1 ajr. • New version of pirate in August CCP 4 helpdesk: ccp 4@dl. ac. uk
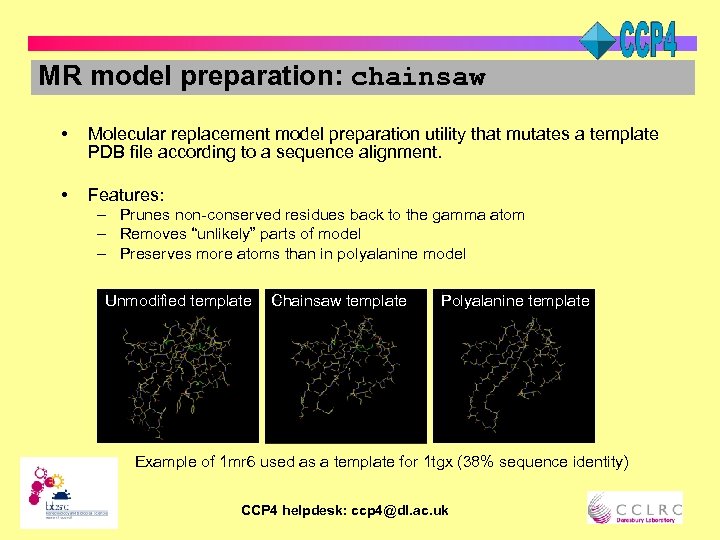 MR model preparation: chainsaw • Molecular replacement model preparation utility that mutates a template PDB file according to a sequence alignment. • Features: – Prunes non-conserved residues back to the gamma atom – Removes “unlikely” parts of model – Preserves more atoms than in polyalanine model Unmodified template Chainsaw template Polyalanine template Example of 1 mr 6 used as a template for 1 tgx (38% sequence identity) CCP 4 helpdesk: ccp 4@dl. ac. uk
MR model preparation: chainsaw • Molecular replacement model preparation utility that mutates a template PDB file according to a sequence alignment. • Features: – Prunes non-conserved residues back to the gamma atom – Removes “unlikely” parts of model – Preserves more atoms than in polyalanine model Unmodified template Chainsaw template Polyalanine template Example of 1 mr 6 used as a template for 1 tgx (38% sequence identity) CCP 4 helpdesk: ccp 4@dl. ac. uk
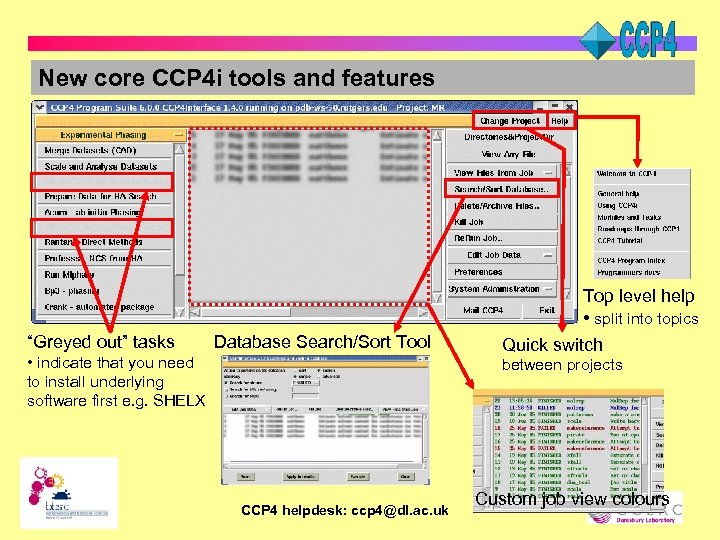 New core CCP 4 i tools and features “Greyed out” tasks Database Search/Sort Tool • indicate that you need to install underlying software first e. g. SHELX Top level help • split into topics Quick switch between projects CCP 4 helpdesk: ccp 4@dl. ac. uk Custom job view colours
New core CCP 4 i tools and features “Greyed out” tasks Database Search/Sort Tool • indicate that you need to install underlying software first e. g. SHELX Top level help • split into topics Quick switch between projects CCP 4 helpdesk: ccp 4@dl. ac. uk Custom job view colours
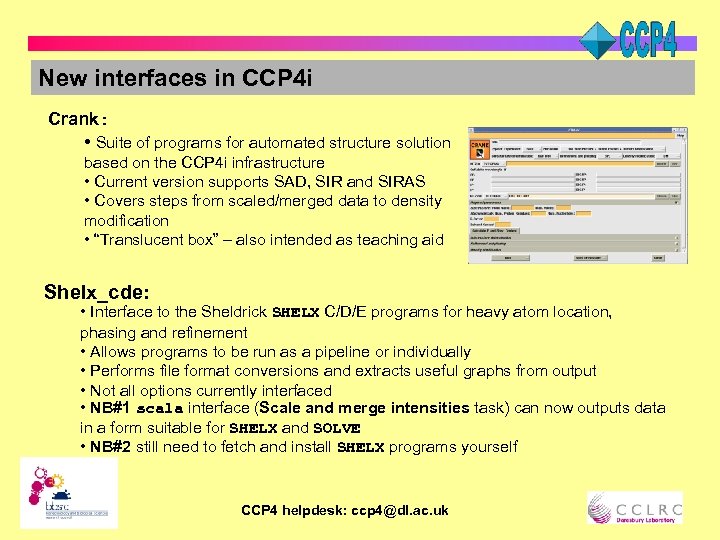 New interfaces in CCP 4 i Crank: • Suite of programs for automated structure solution based on the CCP 4 i infrastructure • Current version supports SAD, SIR and SIRAS • Covers steps from scaled/merged data to density modification • “Translucent box” – also intended as teaching aid Shelx_cde: • Interface to the Sheldrick SHELX C/D/E programs for heavy atom location, phasing and refinement • Allows programs to be run as a pipeline or individually • Performs file format conversions and extracts useful graphs from output • Not all options currently interfaced • NB#1 scala interface (Scale and merge intensities task) can now outputs data in a form suitable for SHELX and SOLVE • NB#2 still need to fetch and install SHELX programs yourself CCP 4 helpdesk: ccp 4@dl. ac. uk
New interfaces in CCP 4 i Crank: • Suite of programs for automated structure solution based on the CCP 4 i infrastructure • Current version supports SAD, SIR and SIRAS • Covers steps from scaled/merged data to density modification • “Translucent box” – also intended as teaching aid Shelx_cde: • Interface to the Sheldrick SHELX C/D/E programs for heavy atom location, phasing and refinement • Allows programs to be run as a pipeline or individually • Performs file format conversions and extracts useful graphs from output • Not all options currently interfaced • NB#1 scala interface (Scale and merge intensities task) can now outputs data in a form suitable for SHELX and SOLVE • NB#2 still need to fetch and install SHELX programs yourself CCP 4 helpdesk: ccp 4@dl. ac. uk
 New/updated interfaces in CCP 4 i Interfaces for new programs: • Pirate and Clipper utilities • BP 3 (in the Experimental Phasing module) • Superpose • Chainsaw (“Create Search Model” in Molecular Replacement module) • Also a launcher task for CCP 4 mg launcher (see later) Plus updates to existing tasks, for example: • areaimol (accessible surface area calculation): generate pairwise area differences for arbitrary groups of chains • matthews_coef and pdbset/pdbcur: accommodate new/extended functionality CCP 4 helpdesk: ccp 4@dl. ac. uk
New/updated interfaces in CCP 4 i Interfaces for new programs: • Pirate and Clipper utilities • BP 3 (in the Experimental Phasing module) • Superpose • Chainsaw (“Create Search Model” in Molecular Replacement module) • Also a launcher task for CCP 4 mg launcher (see later) Plus updates to existing tasks, for example: • areaimol (accessible surface area calculation): generate pairwise area differences for arbitrary groups of chains • matthews_coef and pdbset/pdbcur: accommodate new/extended functionality CCP 4 helpdesk: ccp 4@dl. ac. uk
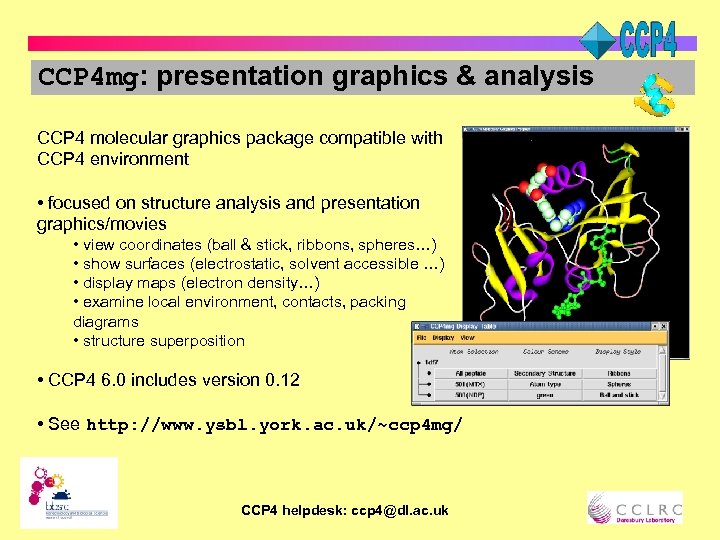 CCP 4 mg: presentation graphics & analysis CCP 4 molecular graphics package compatible with CCP 4 environment • focused on structure analysis and presentation graphics/movies • view coordinates (ball & stick, ribbons, spheres…) • show surfaces (electrostatic, solvent accessible …) • display maps (electron density…) • examine local environment, contacts, packing diagrams • structure superposition • CCP 4 6. 0 includes version 0. 12 • See http: //www. ysbl. york. ac. uk/~ccp 4 mg/ CCP 4 helpdesk: ccp 4@dl. ac. uk
CCP 4 mg: presentation graphics & analysis CCP 4 molecular graphics package compatible with CCP 4 environment • focused on structure analysis and presentation graphics/movies • view coordinates (ball & stick, ribbons, spheres…) • show surfaces (electrostatic, solvent accessible …) • display maps (electron density…) • examine local environment, contacts, packing diagrams • structure superposition • CCP 4 6. 0 includes version 0. 12 • See http: //www. ysbl. york. ac. uk/~ccp 4 mg/ CCP 4 helpdesk: ccp 4@dl. ac. uk
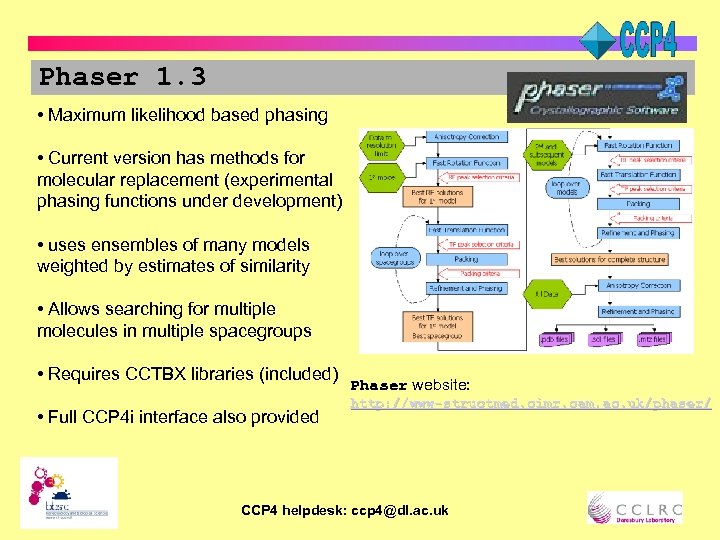 Phaser 1. 3 • Maximum likelihood based phasing • Current version has methods for molecular replacement (experimental phasing functions under development) • uses ensembles of many models weighted by estimates of similarity • Allows searching for multiple molecules in multiple spacegroups • Requires CCTBX libraries (included) • Full CCP 4 i interface also provided Phaser website: http: //www-structmed. cimr. cam. ac. uk/phaser/ CCP 4 helpdesk: ccp 4@dl. ac. uk
Phaser 1. 3 • Maximum likelihood based phasing • Current version has methods for molecular replacement (experimental phasing functions under development) • uses ensembles of many models weighted by estimates of similarity • Allows searching for multiple molecules in multiple spacegroups • Requires CCTBX libraries (included) • Full CCP 4 i interface also provided Phaser website: http: //www-structmed. cimr. cam. ac. uk/phaser/ CCP 4 helpdesk: ccp 4@dl. ac. uk
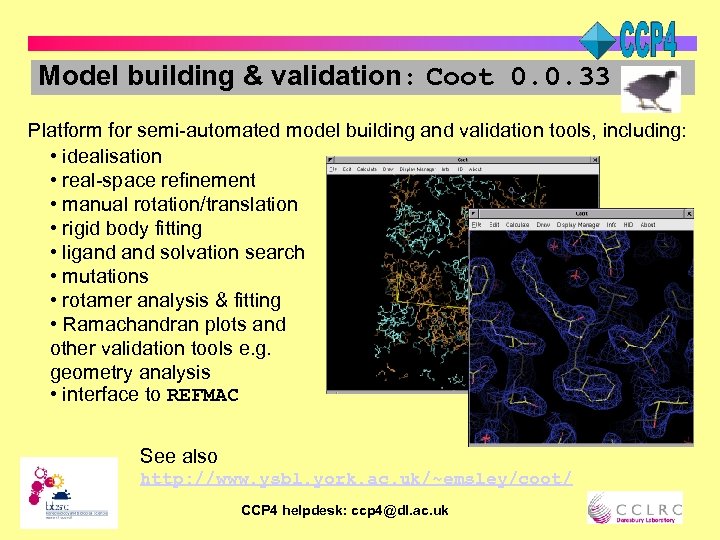 Model building & validation: Coot 0. 0. 33 Platform for semi-automated model building and validation tools, including: • idealisation • real-space refinement • manual rotation/translation • rigid body fitting • ligand solvation search • mutations • rotamer analysis & fitting • Ramachandran plots and other validation tools e. g. geometry analysis • interface to REFMAC See also http: //www. ysbl. york. ac. uk/~emsley/coot/ CCP 4 helpdesk: ccp 4@dl. ac. uk
Model building & validation: Coot 0. 0. 33 Platform for semi-automated model building and validation tools, including: • idealisation • real-space refinement • manual rotation/translation • rigid body fitting • ligand solvation search • mutations • rotamer analysis & fitting • Ramachandran plots and other validation tools e. g. geometry analysis • interface to REFMAC See also http: //www. ysbl. york. ac. uk/~emsley/coot/ CCP 4 helpdesk: ccp 4@dl. ac. uk
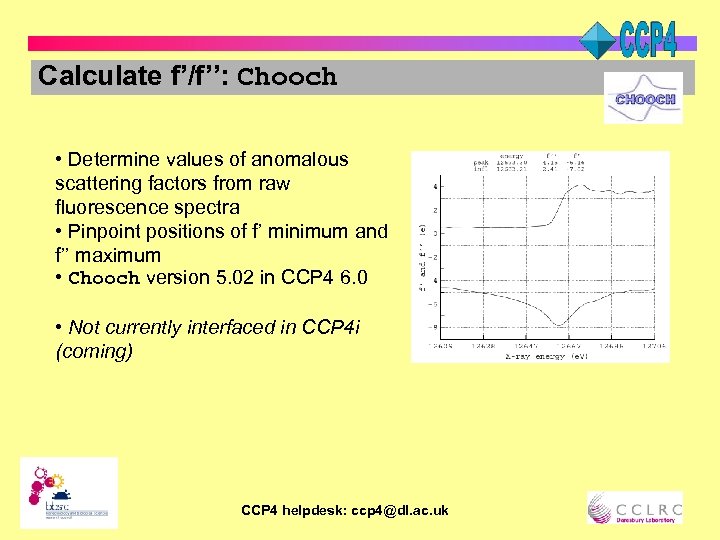 Calculate f’/f’’: Chooch • Determine values of anomalous scattering factors from raw fluorescence spectra • Pinpoint positions of f’ minimum and f’’ maximum • Chooch version 5. 02 in CCP 4 6. 0 • Not currently interfaced in CCP 4 i (coming) CCP 4 helpdesk: ccp 4@dl. ac. uk
Calculate f’/f’’: Chooch • Determine values of anomalous scattering factors from raw fluorescence spectra • Pinpoint positions of f’ minimum and f’’ maximum • Chooch version 5. 02 in CCP 4 6. 0 • Not currently interfaced in CCP 4 i (coming) CCP 4 helpdesk: ccp 4@dl. ac. uk
 Beyond CCP 4 6. 0 CCP 4 helpdesk: ccp 4@dl. ac. uk
Beyond CCP 4 6. 0 CCP 4 helpdesk: ccp 4@dl. ac. uk
 Ongoing developments within CCP 4: programs Available now: • Mr. BUMP: automated Molecular Replacement http: //www. ccp 4. ac. uk/Mr. BUMP • i. MOSFLM: new interface to MOSFLM Other on-going developments within core group: • HAPPy (Heavy Atom Phasing in Python): • an automated Experimental Phasing pipeline • CCP 4 i/BIOXHIT project database: • expanding the data stored by CCP 4 i and making it available to other programs • XIA-DP: • an automated data processing system CCP 4 helpdesk: ccp 4@dl. ac. uk
Ongoing developments within CCP 4: programs Available now: • Mr. BUMP: automated Molecular Replacement http: //www. ccp 4. ac. uk/Mr. BUMP • i. MOSFLM: new interface to MOSFLM Other on-going developments within core group: • HAPPy (Heavy Atom Phasing in Python): • an automated Experimental Phasing pipeline • CCP 4 i/BIOXHIT project database: • expanding the data stored by CCP 4 i and making it available to other programs • XIA-DP: • an automated data processing system CCP 4 helpdesk: ccp 4@dl. ac. uk
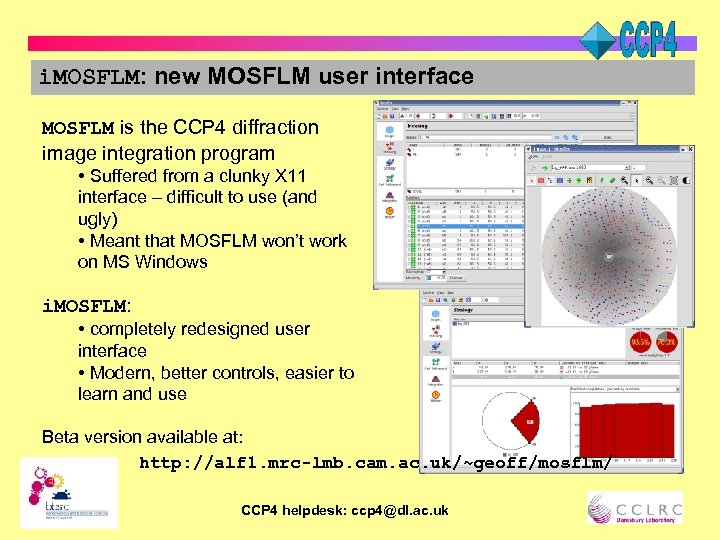 i. MOSFLM: new MOSFLM user interface MOSFLM is the CCP 4 diffraction image integration program • Suffered from a clunky X 11 interface – difficult to use (and ugly) • Meant that MOSFLM won’t work on MS Windows i. MOSFLM: • completely redesigned user interface • Modern, better controls, easier to learn and use Beta version available at: http: //alf 1. mrc-lmb. cam. ac. uk/~geoff/mosflm/ CCP 4 helpdesk: ccp 4@dl. ac. uk
i. MOSFLM: new MOSFLM user interface MOSFLM is the CCP 4 diffraction image integration program • Suffered from a clunky X 11 interface – difficult to use (and ugly) • Meant that MOSFLM won’t work on MS Windows i. MOSFLM: • completely redesigned user interface • Modern, better controls, easier to learn and use Beta version available at: http: //alf 1. mrc-lmb. cam. ac. uk/~geoff/mosflm/ CCP 4 helpdesk: ccp 4@dl. ac. uk
 Some future programs More Clipper-based programs (Kevin Cowtan): • Buccaneer: statistical chain-tracing with ML target • Privateer: refine crude low-resolution structures • See http: //www. ysbl. york. ac. uk/~cowtan/ Pointless: • Spacegroup determination/(re)indexing for unmerged reflection data (Phil Evans) Othercell: • For given cell and lattice type, list alternative indexing and/or cell parameters (Phil Evans) BALBES: • Automated MR using REFMAC, MOLREP and SFCHECK (Garib Murshudov, Alexei Vagin and Fei Long) CCP 4 helpdesk: ccp 4@dl. ac. uk
Some future programs More Clipper-based programs (Kevin Cowtan): • Buccaneer: statistical chain-tracing with ML target • Privateer: refine crude low-resolution structures • See http: //www. ysbl. york. ac. uk/~cowtan/ Pointless: • Spacegroup determination/(re)indexing for unmerged reflection data (Phil Evans) Othercell: • For given cell and lattice type, list alternative indexing and/or cell parameters (Phil Evans) BALBES: • Automated MR using REFMAC, MOLREP and SFCHECK (Garib Murshudov, Alexei Vagin and Fei Long) CCP 4 helpdesk: ccp 4@dl. ac. uk
 TLS Refinement CCP 4 helpdesk: ccp 4@dl. ac. uk
TLS Refinement CCP 4 helpdesk: ccp 4@dl. ac. uk
 TLS Refinement TLS parameterization allows to partly take into account anisotropic motions at medium/low resolution (> 3. 5 Å) ● TLS refinement might improve refinement statistics of several percent ● TLS refinement in REFMAC 5 is fast and therefore can be used routinely ● TLS parameters can be analyzed to extract physical significance ● CCP 4 helpdesk: ccp 4@dl. ac. uk
TLS Refinement TLS parameterization allows to partly take into account anisotropic motions at medium/low resolution (> 3. 5 Å) ● TLS refinement might improve refinement statistics of several percent ● TLS refinement in REFMAC 5 is fast and therefore can be used routinely ● TLS parameters can be analyzed to extract physical significance ● CCP 4 helpdesk: ccp 4@dl. ac. uk
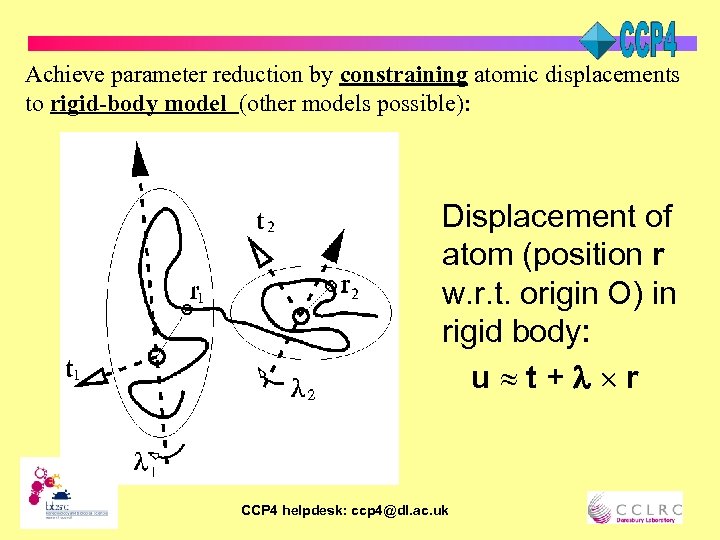 Achieve parameter reduction by constraining atomic displacements to rigid-body model (other models possible): Displacement of atom (position r w. r. t. origin O) in rigid body: u t+ r CCP 4 helpdesk: ccp 4@dl. ac. uk
Achieve parameter reduction by constraining atomic displacements to rigid-body model (other models possible): Displacement of atom (position r w. r. t. origin O) in rigid body: u t+ r CCP 4 helpdesk: ccp 4@dl. ac. uk
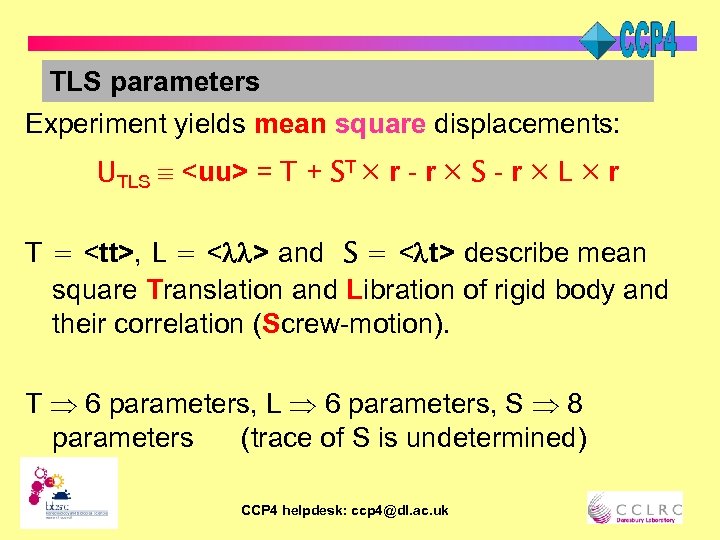 TLS parameters Experiment yields mean square displacements: UTLS
TLS parameters Experiment yields mean square displacements: UTLS
 Atomic parameters from TLSANL • TLS tensors UTLS for atoms in group (ANISOU records in XYZOUT) • UTLS BTLS and anisotropy A • Also have individually refined Bres from Refmac • BTOT = BTLS + Bres (ATOM records in XYZOUT) • keyword ISOOUT determines which contribution recorded in ATOM/ANISOU lines CCP 4 helpdesk: ccp 4@dl. ac. uk
Atomic parameters from TLSANL • TLS tensors UTLS for atoms in group (ANISOU records in XYZOUT) • UTLS BTLS and anisotropy A • Also have individually refined Bres from Refmac • BTOT = BTLS + Bres (ATOM records in XYZOUT) • keyword ISOOUT determines which contribution recorded in ATOM/ANISOU lines CCP 4 helpdesk: ccp 4@dl. ac. uk


