f03a35176f8e81d3665ad23e2fa5fec1.ppt
- Количество слайдов: 25
 What drives antigenic drift in a single influenza season? Maciej F. Boni Stanford University, Department of Biological Sciences Co-authors: Julia R. Gog, Viggo Andreasen, Marcus W. Feldman DIMACS Workshop on the Epidemiology and Evolution of Influenza Rutgers University, January 26, 2006
What drives antigenic drift in a single influenza season? Maciej F. Boni Stanford University, Department of Biological Sciences Co-authors: Julia R. Gog, Viggo Andreasen, Marcus W. Feldman DIMACS Workshop on the Epidemiology and Evolution of Influenza Rutgers University, January 26, 2006
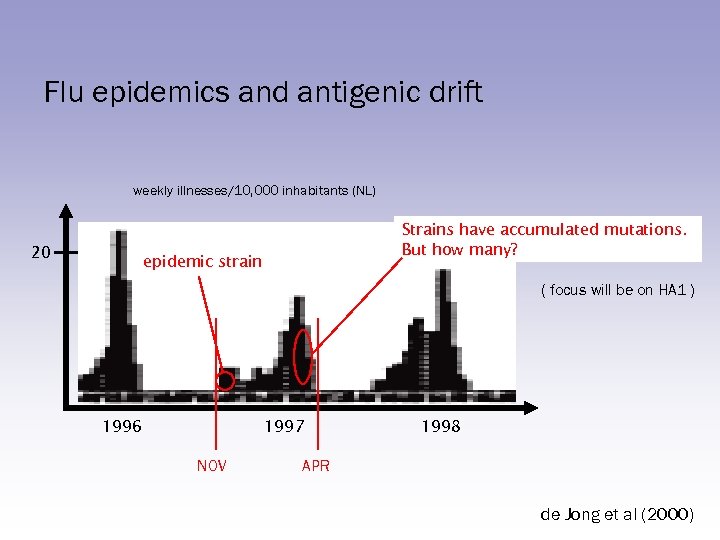 Flu epidemics and antigenic drift weekly illnesses/10, 000 inhabitants (NL) 20 Strains have accumulated mutations. But how many? epidemic strain ( focus will be on HA 1 ) 1996 1997 NOV 1998 APR de Jong et al (2000)
Flu epidemics and antigenic drift weekly illnesses/10, 000 inhabitants (NL) 20 Strains have accumulated mutations. But how many? epidemic strain ( focus will be on HA 1 ) 1996 1997 NOV 1998 APR de Jong et al (2000)
 HA 1 polymorphism – local datasets n Coiras et al, Arch. Vir. (2001) n Schweiger et al, Med. Microbiol. Immunol. (2002) n Pyhälä et al, J. Med. Virol. (2004) mean within-season distance = 2. 8 aa (6 nt) max within-season distance = 8 aa (25 nt)
HA 1 polymorphism – local datasets n Coiras et al, Arch. Vir. (2001) n Schweiger et al, Med. Microbiol. Immunol. (2002) n Pyhälä et al, J. Med. Virol. (2004) mean within-season distance = 2. 8 aa (6 nt) max within-season distance = 8 aa (25 nt)
 Neutral Epidemic Model Number of infections with epidemic-originating strain Number of infections with a strain k mutations away
Neutral Epidemic Model Number of infections with epidemic-originating strain Number of infections with a strain k mutations away
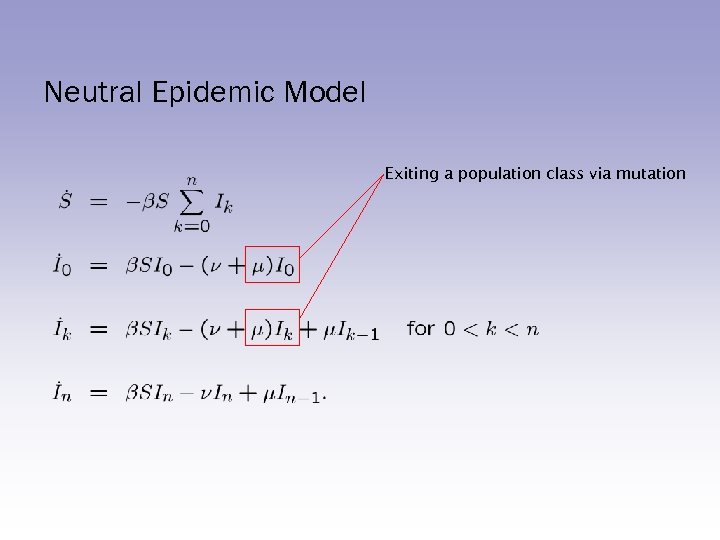 Neutral Epidemic Model Exiting a population class via mutation
Neutral Epidemic Model Exiting a population class via mutation
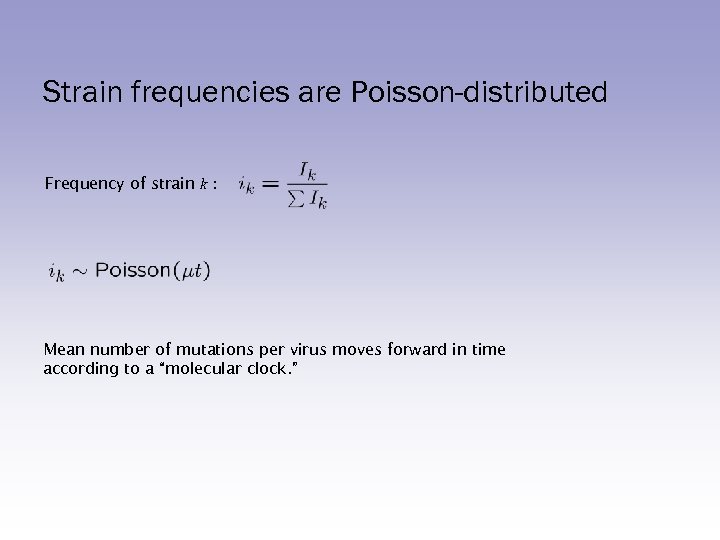 Strain frequencies are Poisson-distributed Frequency of strain k : Mean number of mutations per virus moves forward in time according to a “molecular clock. ”
Strain frequencies are Poisson-distributed Frequency of strain k : Mean number of mutations per virus moves forward in time according to a “molecular clock. ”
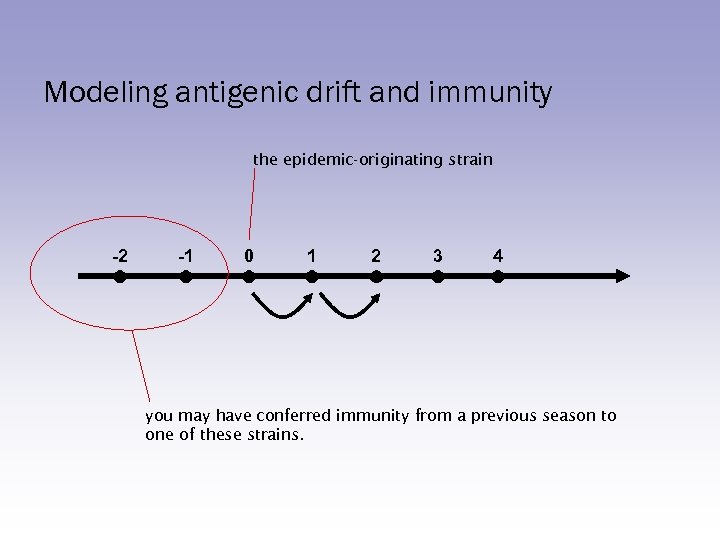 Modeling antigenic drift and immunity the epidemic-originating strain -2 -1 0 1 2 3 4 you may have conferred immunity from a previous season to one of these strains.
Modeling antigenic drift and immunity the epidemic-originating strain -2 -1 0 1 2 3 4 you may have conferred immunity from a previous season to one of these strains.
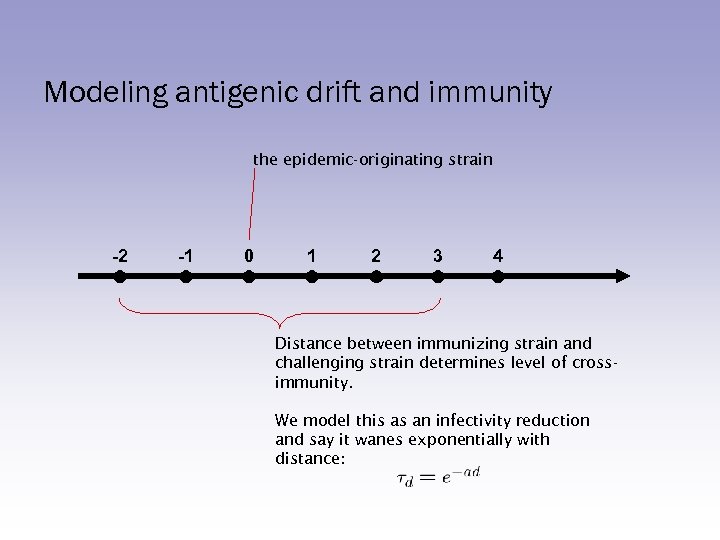 Modeling antigenic drift and immunity the epidemic-originating strain -2 -1 0 1 2 3 4 Distance between immunizing strain and challenging strain determines level of crossimmunity. We model this as an infectivity reduction and say it wanes exponentially with distance:
Modeling antigenic drift and immunity the epidemic-originating strain -2 -1 0 1 2 3 4 Distance between immunizing strain and challenging strain determines level of crossimmunity. We model this as an infectivity reduction and say it wanes exponentially with distance:
 Non-neutral model n n Amino-acid replacements in influenza surface proteins confer a fitness benefit via increased transmissibility Hosts have some immunity structure from vaccination or previous infections ( need both )
Non-neutral model n n Amino-acid replacements in influenza surface proteins confer a fitness benefit via increased transmissibility Hosts have some immunity structure from vaccination or previous infections ( need both )
 Keeping track of hosts q 0 q 30 completely immune ( to I 0 ) completely naive j+k is distance between immunizing and challenging (infecting) strain
Keeping track of hosts q 0 q 30 completely immune ( to I 0 ) completely naive j+k is distance between immunizing and challenging (infecting) strain
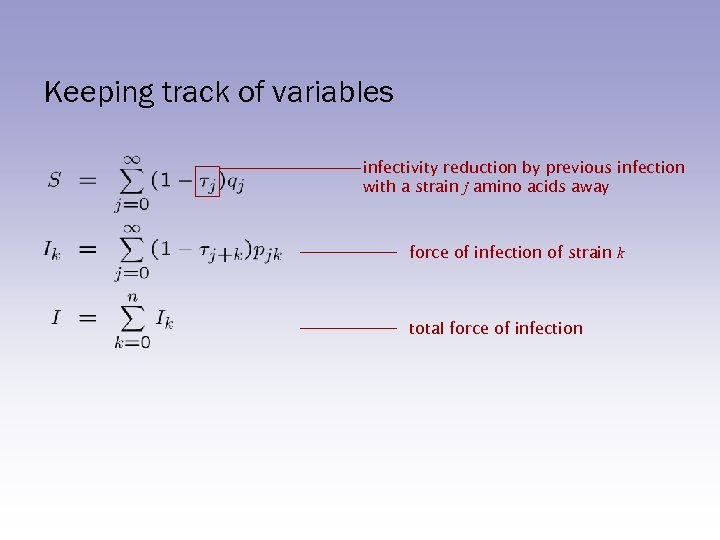 Keeping track of variables infectivity reduction by previous infection with a strain j amino acids away force of infection of strain k total force of infection
Keeping track of variables infectivity reduction by previous infection with a strain j amino acids away force of infection of strain k total force of infection
 Equations
Equations
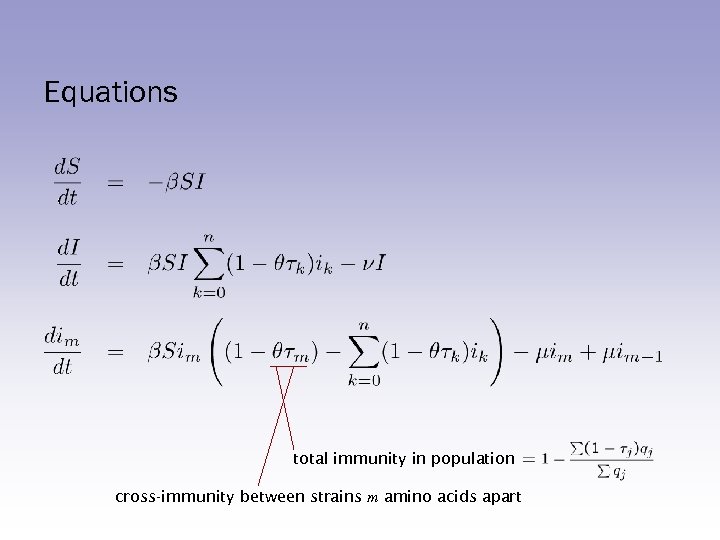 Equations total immunity in population cross-immunity between strains m amino acids apart
Equations total immunity in population cross-immunity between strains m amino acids apart
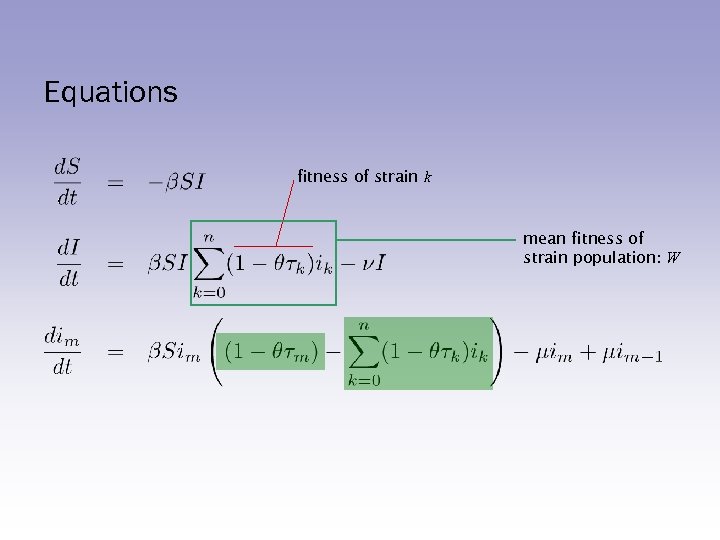 Equations fitness of strain k mean fitness of strain population: W
Equations fitness of strain k mean fitness of strain population: W
 Population genetics Fisher’s Fundamental Theorem Define mean antigenic drift in virus population as: This is the Price Equation, thus, the basic influenza population dynamics can be viewed in a standard population genetic framework.
Population genetics Fisher’s Fundamental Theorem Define mean antigenic drift in virus population as: This is the Price Equation, thus, the basic influenza population dynamics can be viewed in a standard population genetic framework.
 Under neutrality
Under neutrality
 I(t) Takes 7 aa-changes to escape 50% immunity
I(t) Takes 7 aa-changes to escape 50% immunity
 Define the excess antigenic drift as: How do you know when the epidemic ends?
Define the excess antigenic drift as: How do you know when the epidemic ends?
 I(t)
I(t)
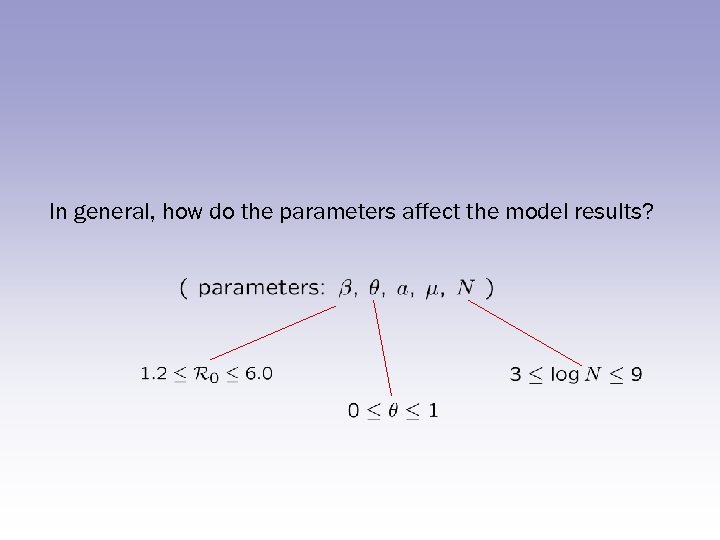 In general, how do the parameters affect the model results?
In general, how do the parameters affect the model results?
 Partial correlations immunity : immune-escape/mutation :
Partial correlations immunity : immune-escape/mutation :
 Partial correlations immunity : immune-escape/mutation :
Partial correlations immunity : immune-escape/mutation :
 Host immunity drives antigenic drift
Host immunity drives antigenic drift
 Public health implications n n Vaccination strategies: under-vaccination or imperfect vaccination may cause much excess antigenic drift. Pandemic implications: need to consider the effects of vaccination during the 2 nd year after a pandemic, and their effects on the 3 rd year after a pandemic.
Public health implications n n Vaccination strategies: under-vaccination or imperfect vaccination may cause much excess antigenic drift. Pandemic implications: need to consider the effects of vaccination during the 2 nd year after a pandemic, and their effects on the 3 rd year after a pandemic.
 Thanks Viggo Andreasen University of Roskile, Department of Mathematics and Physics Julia Gog Cambridge University, Department of Zoology Marc Feldman Stanford University, Department of Biological Sciences Freddy Christiansen University of Aarhus, Department of Biology Mike Macpherson Stanford University, Department of Biological Sciences ( and for funding to NIH grant GM 28016, NSF, and Stanford University )
Thanks Viggo Andreasen University of Roskile, Department of Mathematics and Physics Julia Gog Cambridge University, Department of Zoology Marc Feldman Stanford University, Department of Biological Sciences Freddy Christiansen University of Aarhus, Department of Biology Mike Macpherson Stanford University, Department of Biological Sciences ( and for funding to NIH grant GM 28016, NSF, and Stanford University )


