4bfa07e884cc2a616a8578d8a5df5087.ppt
- Количество слайдов: 21
 We have alredy discussed Ø What is MSA (Multiple Sequence Alignment)? • What is it good for? • How do I use it? Ø Software and algorithms • The programs • How they work? • Which to use? Ø In practice • Get the sequences • Reformat them • Evaluate the alignment • Realign or modify the alignment • Add or subtract sequence Lec 09
We have alredy discussed Ø What is MSA (Multiple Sequence Alignment)? • What is it good for? • How do I use it? Ø Software and algorithms • The programs • How they work? • Which to use? Ø In practice • Get the sequences • Reformat them • Evaluate the alignment • Realign or modify the alignment • Add or subtract sequence Lec 09
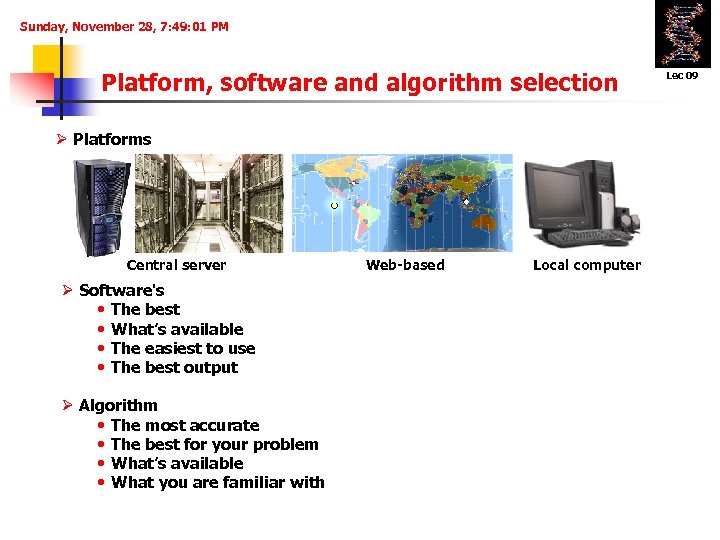 Sunday, November 28, 7: 49: 01 PM Platform, software and algorithm selection Ø Platforms Central server Ø Software's • The best • What’s available • The easiest to use • The best output Ø Algorithm • The most accurate • The best for your problem • What’s available • What you are familiar with Web-based Local computer Lec 09
Sunday, November 28, 7: 49: 01 PM Platform, software and algorithm selection Ø Platforms Central server Ø Software's • The best • What’s available • The easiest to use • The best output Ø Algorithm • The most accurate • The best for your problem • What’s available • What you are familiar with Web-based Local computer Lec 09
 Sunday, November 28, 7: 49: 01 PM Main applications of MSA Lec 09
Sunday, November 28, 7: 49: 01 PM Main applications of MSA Lec 09
 Sunday, November 28, 7: 49: 01 PM Get the sequences: databases Lec 09 • Gen. Bank: An annotated collection of all publicly available nucleotide and protein sequences. • Ref. Seq: NCBI non-redundant set of reference sequences, including genomic DNA, transcript (RNA), and protein products. • Uni. Prot Consortium Database: Universal protein knowledgebase, a central resource of protein sequence and function from Swiss-Prot, Tr. EMBL and PIR. • Entrez Gene: Gene-centered information at NCBI. • Uni. Gene: Unified clusters of ESTs and full-length m. RNA sequences. • OMIM: Online Mendelian inheritance in man: a catalog of human genetic and genomic disorders. • Model Organism Genome Databases: MGD, RGD, SGD, Flybase… • Gene. Cards: Integrated database of human genes, maps, proteins and diseases. • SNP Consortium Database.
Sunday, November 28, 7: 49: 01 PM Get the sequences: databases Lec 09 • Gen. Bank: An annotated collection of all publicly available nucleotide and protein sequences. • Ref. Seq: NCBI non-redundant set of reference sequences, including genomic DNA, transcript (RNA), and protein products. • Uni. Prot Consortium Database: Universal protein knowledgebase, a central resource of protein sequence and function from Swiss-Prot, Tr. EMBL and PIR. • Entrez Gene: Gene-centered information at NCBI. • Uni. Gene: Unified clusters of ESTs and full-length m. RNA sequences. • OMIM: Online Mendelian inheritance in man: a catalog of human genetic and genomic disorders. • Model Organism Genome Databases: MGD, RGD, SGD, Flybase… • Gene. Cards: Integrated database of human genes, maps, proteins and diseases. • SNP Consortium Database.
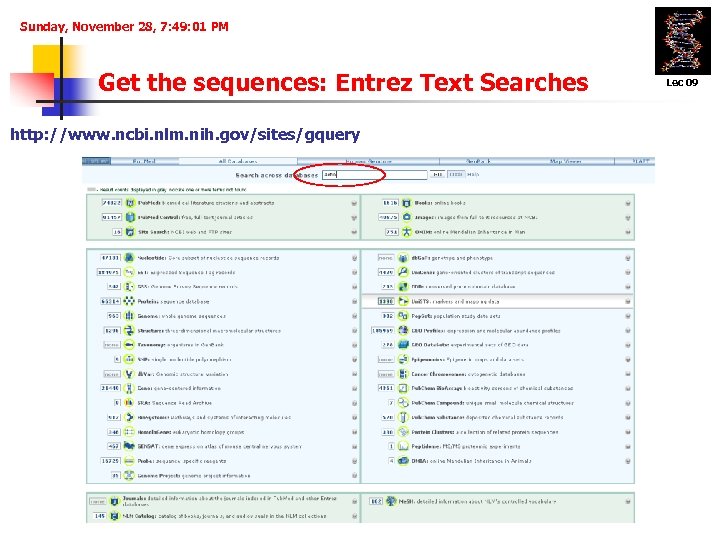 Sunday, November 28, 7: 49: 01 PM Get the sequences: Entrez Text Searches http: //www. ncbi. nlm. nih. gov/sites/gquery Lec 09
Sunday, November 28, 7: 49: 01 PM Get the sequences: Entrez Text Searches http: //www. ncbi. nlm. nih. gov/sites/gquery Lec 09
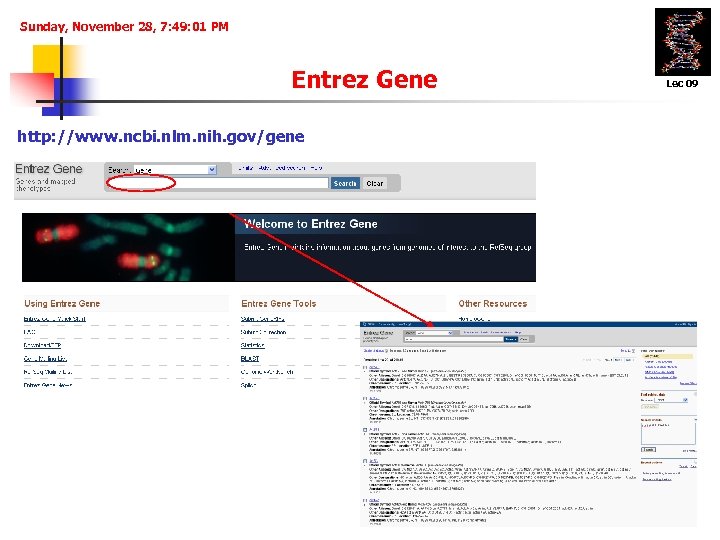 Sunday, November 28, 7: 49: 01 PM Entrez Gene http: //www. ncbi. nlm. nih. gov/gene Lec 09
Sunday, November 28, 7: 49: 01 PM Entrez Gene http: //www. ncbi. nlm. nih. gov/gene Lec 09
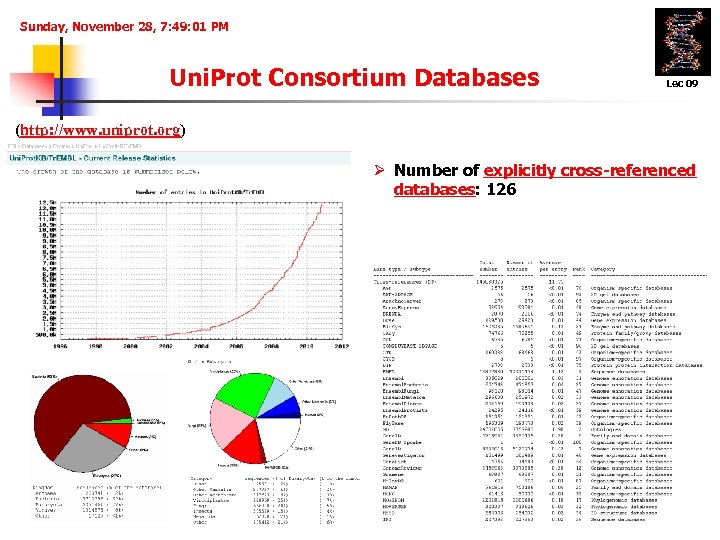 Sunday, November 28, 7: 49: 01 PM Uni. Prot Consortium Databases Lec 09 (http: //www. uniprot. org) Ø Number of explicitly cross-referenced databases: 126
Sunday, November 28, 7: 49: 01 PM Uni. Prot Consortium Databases Lec 09 (http: //www. uniprot. org) Ø Number of explicitly cross-referenced databases: 126
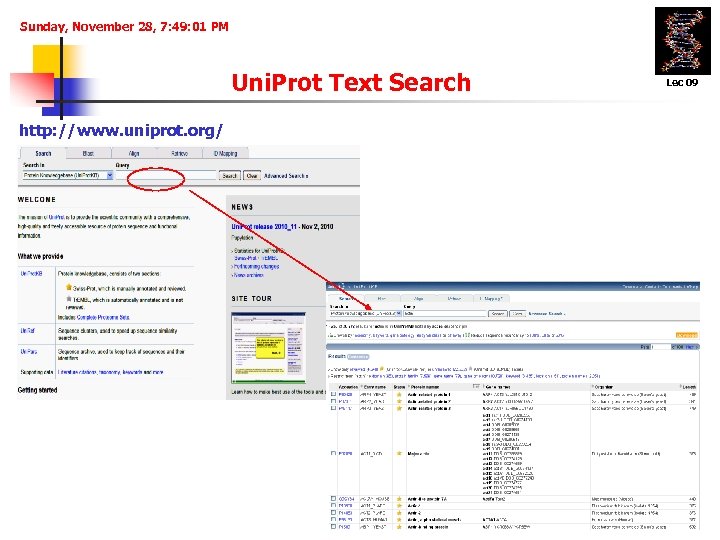 Sunday, November 28, 7: 49: 01 PM Uni. Prot Text Search http: //www. uniprot. org/ Lec 09
Sunday, November 28, 7: 49: 01 PM Uni. Prot Text Search http: //www. uniprot. org/ Lec 09
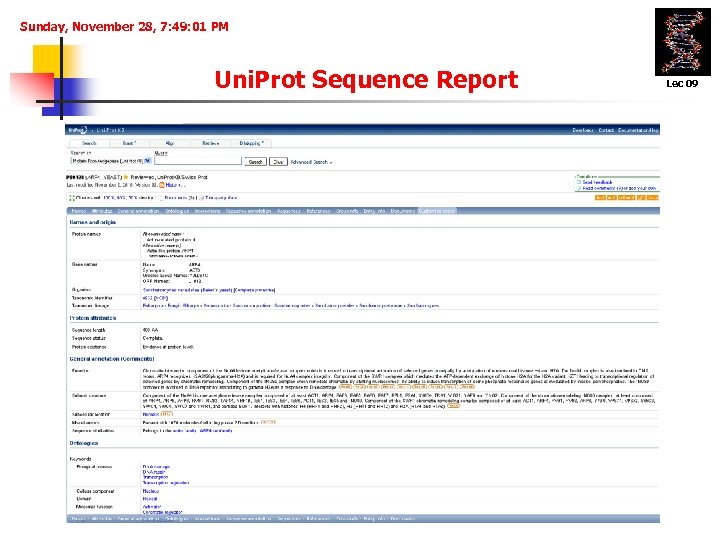 Sunday, November 28, 7: 49: 01 PM Uni. Prot Sequence Report Lec 09
Sunday, November 28, 7: 49: 01 PM Uni. Prot Sequence Report Lec 09
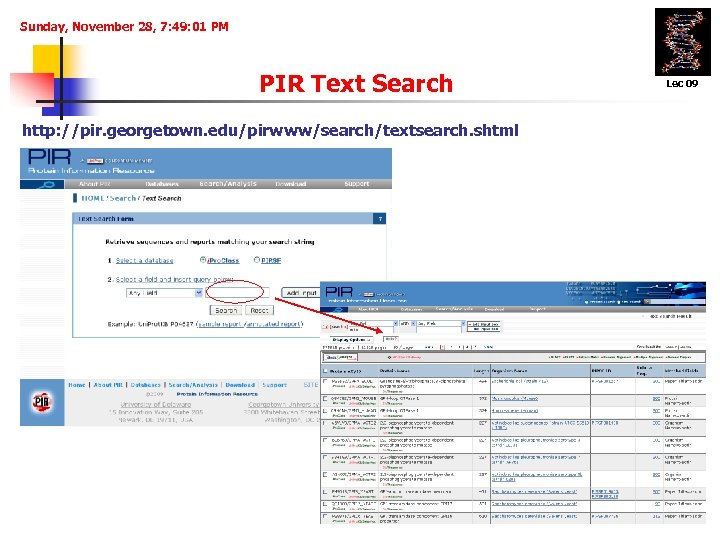 Sunday, November 28, 7: 49: 01 PM PIR Text Search http: //pir. georgetown. edu/pirwww/search/textsearch. shtml Lec 09
Sunday, November 28, 7: 49: 01 PM PIR Text Search http: //pir. georgetown. edu/pirwww/search/textsearch. shtml Lec 09
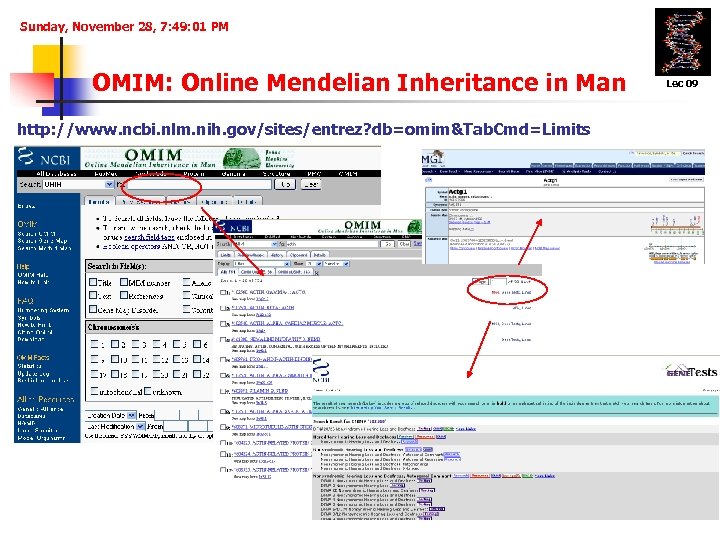 Sunday, November 28, 7: 49: 01 PM OMIM: Online Mendelian Inheritance in Man http: //www. ncbi. nlm. nih. gov/sites/entrez? db=omim&Tab. Cmd=Limits Lec 09
Sunday, November 28, 7: 49: 01 PM OMIM: Online Mendelian Inheritance in Man http: //www. ncbi. nlm. nih. gov/sites/entrez? db=omim&Tab. Cmd=Limits Lec 09
 Sunday, November 28, 7: 49: 01 PM Protein Family Databases Lec 09 Ø Whole Proteins • PIRSF: A Network Classification System of Protein Families • COG (Clusters of Orthologous Groups) of Complete Genomes • Proto. Net: Automated Hierarchical Classification of Proteins Ø Protein Domains • Pfam: Alignments and HMM Models of Protein Domains • SMART: Protein Domain Families • CDD: Conserved Domain Database Ø Protein Motifs • PROSITE: Protein Patterns and Profiles • BLOCKS: Protein Sequence Motifs and Alignments • PRINTS: Protein Sequence Motifs and Signatures Ø Integrated Family Databases • i. Pro. Class: Superfamilies/Families, Domains, Motifs, Rich Links • Inter. Pro: Integrate Pfam, PRINTS, PROSITES, Pro. Dom, SMART, PIRSF, Super. Family
Sunday, November 28, 7: 49: 01 PM Protein Family Databases Lec 09 Ø Whole Proteins • PIRSF: A Network Classification System of Protein Families • COG (Clusters of Orthologous Groups) of Complete Genomes • Proto. Net: Automated Hierarchical Classification of Proteins Ø Protein Domains • Pfam: Alignments and HMM Models of Protein Domains • SMART: Protein Domain Families • CDD: Conserved Domain Database Ø Protein Motifs • PROSITE: Protein Patterns and Profiles • BLOCKS: Protein Sequence Motifs and Alignments • PRINTS: Protein Sequence Motifs and Signatures Ø Integrated Family Databases • i. Pro. Class: Superfamilies/Families, Domains, Motifs, Rich Links • Inter. Pro: Integrate Pfam, PRINTS, PROSITES, Pro. Dom, SMART, PIRSF, Super. Family
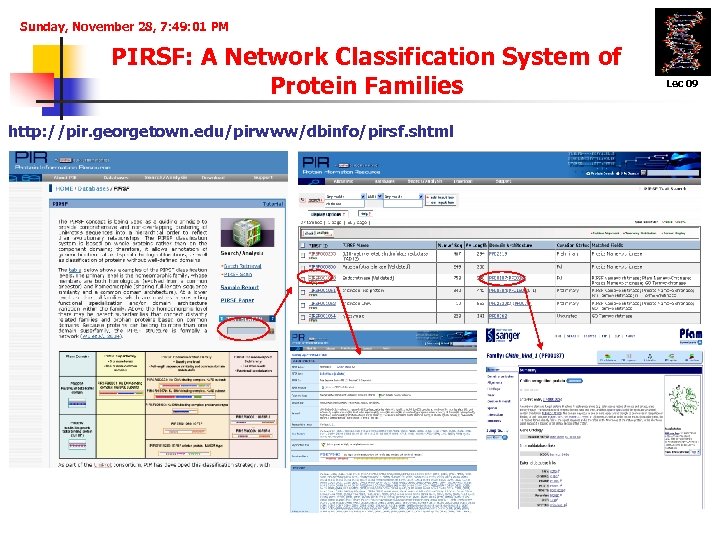 Sunday, November 28, 7: 49: 01 PM PIRSF: A Network Classification System of Protein Families http: //pir. georgetown. edu/pirwww/dbinfo/pirsf. shtml Lec 09
Sunday, November 28, 7: 49: 01 PM PIRSF: A Network Classification System of Protein Families http: //pir. georgetown. edu/pirwww/dbinfo/pirsf. shtml Lec 09
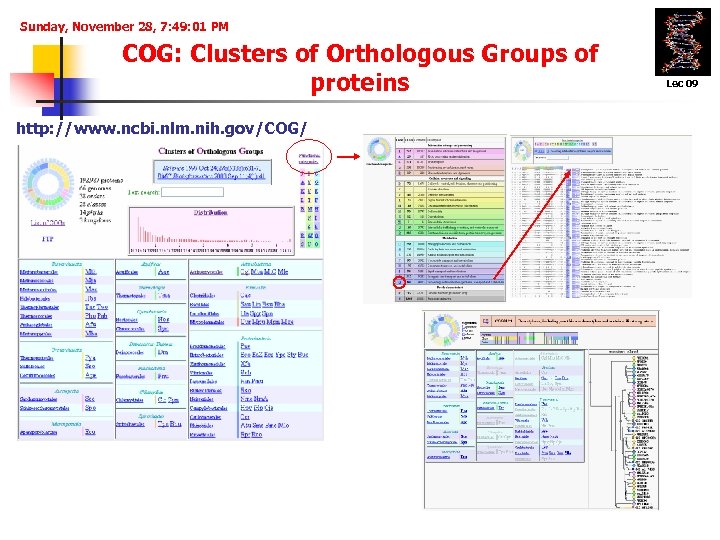 Sunday, November 28, 7: 49: 01 PM COG: Clusters of Orthologous Groups of proteins http: //www. ncbi. nlm. nih. gov/COG/ Lec 09
Sunday, November 28, 7: 49: 01 PM COG: Clusters of Orthologous Groups of proteins http: //www. ncbi. nlm. nih. gov/COG/ Lec 09
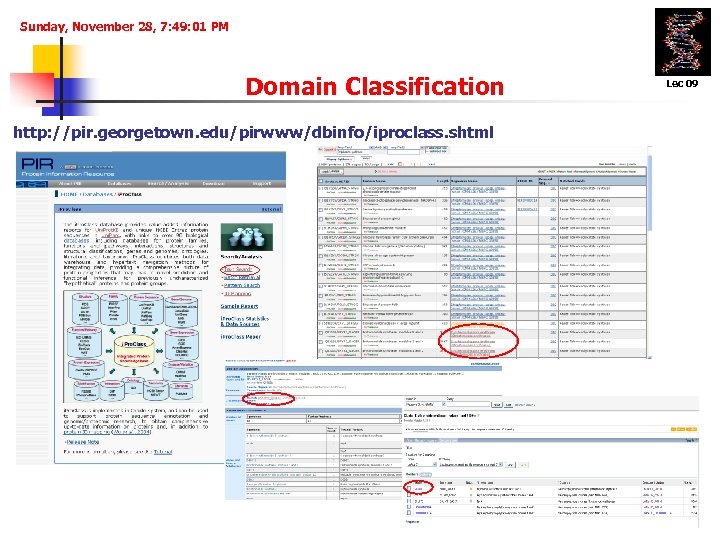 Sunday, November 28, 7: 49: 01 PM Domain Classification http: //pir. georgetown. edu/pirwww/dbinfo/iproclass. shtml Lec 09
Sunday, November 28, 7: 49: 01 PM Domain Classification http: //pir. georgetown. edu/pirwww/dbinfo/iproclass. shtml Lec 09
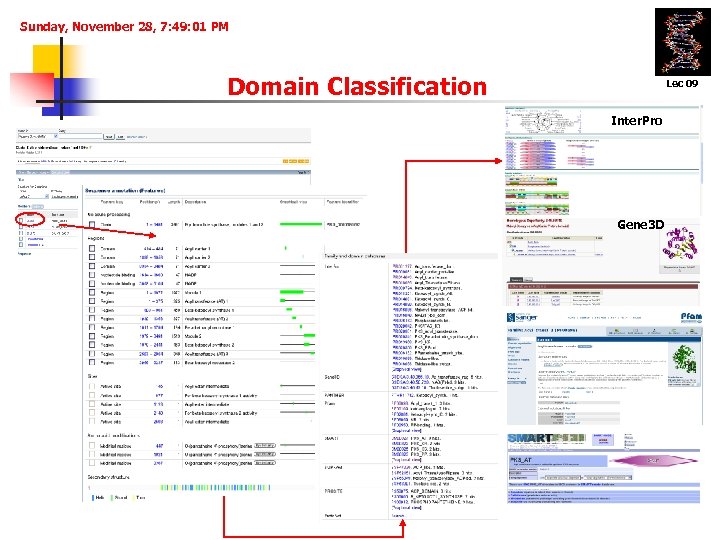 Sunday, November 28, 7: 49: 01 PM Domain Classification Lec 09 Inter. Pro Gene 3 D
Sunday, November 28, 7: 49: 01 PM Domain Classification Lec 09 Inter. Pro Gene 3 D
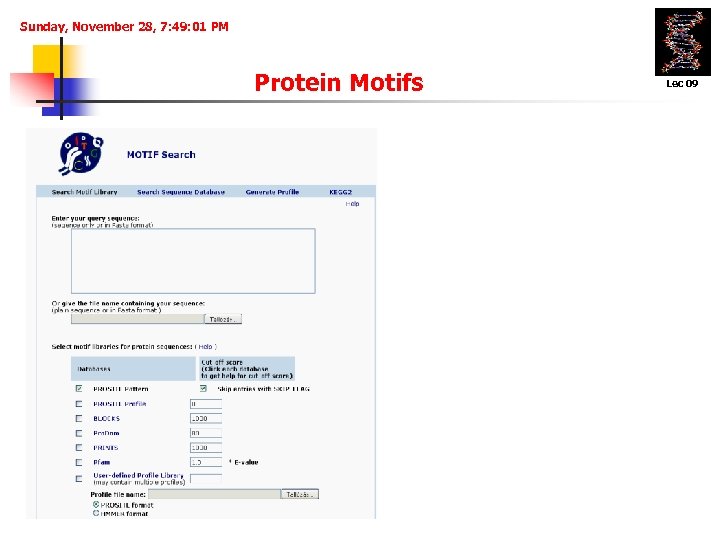 Sunday, November 28, 7: 49: 01 PM Protein Motifs Lec 09
Sunday, November 28, 7: 49: 01 PM Protein Motifs Lec 09
 Sunday, November 28, 7: 49: 01 PM Databases of Protein Functions Lec 09 Ø Metabolic Pathways, Enzymes, and Compounds • Enzyme Classification: Classification and Nomenclature of Enzyme-Catalysed Reactions (EC-IUBMB) • KEGG (Kyoto Encyclopedia of Genes and Genomes): Metabolic Pathways • LIGAND (at KEGG): Chemical Compounds, Reactions and Enzymes • Eco. Cyc: Encyclopedia of E. coli Genes and Metabolism • Meta. Cyc: Metabolic Encyclopedia (Metabolic Pathways) • BRENDA: Enzyme Database • UM-BBD: Microbial Biocatalytic Reactions and Biodegradation Pathways Ø Cellular Regulation and Gene Networks n Epo. DB: Genes Expressed during Human Erythropoiesis n BIND: Descriptions of interactions, molecular complexes and pathways n DIP: Catalogs experimentally determined interactions between proteins n Bio. Carta: Biological pathways of human and mouse n GO: Gene Ontology Consortium Database
Sunday, November 28, 7: 49: 01 PM Databases of Protein Functions Lec 09 Ø Metabolic Pathways, Enzymes, and Compounds • Enzyme Classification: Classification and Nomenclature of Enzyme-Catalysed Reactions (EC-IUBMB) • KEGG (Kyoto Encyclopedia of Genes and Genomes): Metabolic Pathways • LIGAND (at KEGG): Chemical Compounds, Reactions and Enzymes • Eco. Cyc: Encyclopedia of E. coli Genes and Metabolism • Meta. Cyc: Metabolic Encyclopedia (Metabolic Pathways) • BRENDA: Enzyme Database • UM-BBD: Microbial Biocatalytic Reactions and Biodegradation Pathways Ø Cellular Regulation and Gene Networks n Epo. DB: Genes Expressed during Human Erythropoiesis n BIND: Descriptions of interactions, molecular complexes and pathways n DIP: Catalogs experimentally determined interactions between proteins n Bio. Carta: Biological pathways of human and mouse n GO: Gene Ontology Consortium Database
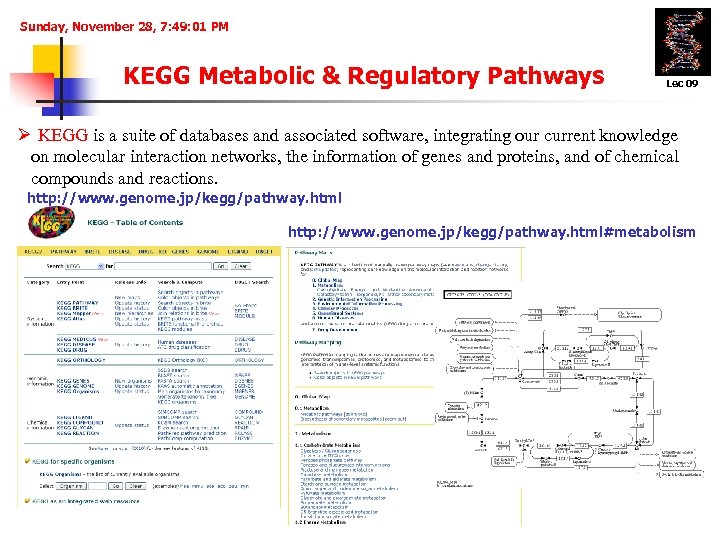 Sunday, November 28, 7: 49: 01 PM KEGG Metabolic & Regulatory Pathways Lec 09 Ø KEGG is a suite of databases and associated software, integrating our current knowledge on molecular interaction networks, the information of genes and proteins, and of chemical compounds and reactions. http: //www. genome. jp/kegg/pathway. html#metabolism
Sunday, November 28, 7: 49: 01 PM KEGG Metabolic & Regulatory Pathways Lec 09 Ø KEGG is a suite of databases and associated software, integrating our current knowledge on molecular interaction networks, the information of genes and proteins, and of chemical compounds and reactions. http: //www. genome. jp/kegg/pathway. html#metabolism
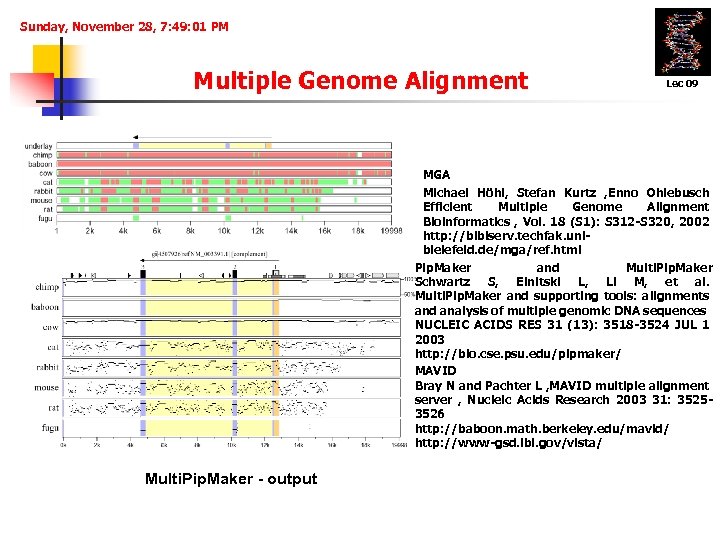 Sunday, November 28, 7: 49: 01 PM Multiple Genome Alignment Lec 09 MGA Michael Höhl, Stefan Kurtz , Enno Ohlebusch Efficient Multiple Genome Alignment Bioinformatics , Vol. 18 (S 1): S 312 -S 320, 2002 http: //bibiserv. techfak. unibielefeld. de/mga/ref. html Pip. Maker and Multi. Pip. Maker Schwartz S, Elnitski L, Li M, et al. Multi. Pip. Maker and supporting tools: alignments and analysis of multiple genomic DNA sequences NUCLEIC ACIDS RES 31 (13): 3518 -3524 JUL 1 2003 http: //bio. cse. psu. edu/pipmaker/ MAVID Bray N and Pachter L , MAVID multiple alignment server , Nucleic Acids Research 2003 31: 35253526 http: //baboon. math. berkeley. edu/mavid/ http: //www-gsd. lbl. gov/vista/ Multi. Pip. Maker - output
Sunday, November 28, 7: 49: 01 PM Multiple Genome Alignment Lec 09 MGA Michael Höhl, Stefan Kurtz , Enno Ohlebusch Efficient Multiple Genome Alignment Bioinformatics , Vol. 18 (S 1): S 312 -S 320, 2002 http: //bibiserv. techfak. unibielefeld. de/mga/ref. html Pip. Maker and Multi. Pip. Maker Schwartz S, Elnitski L, Li M, et al. Multi. Pip. Maker and supporting tools: alignments and analysis of multiple genomic DNA sequences NUCLEIC ACIDS RES 31 (13): 3518 -3524 JUL 1 2003 http: //bio. cse. psu. edu/pipmaker/ MAVID Bray N and Pachter L , MAVID multiple alignment server , Nucleic Acids Research 2003 31: 35253526 http: //baboon. math. berkeley. edu/mavid/ http: //www-gsd. lbl. gov/vista/ Multi. Pip. Maker - output
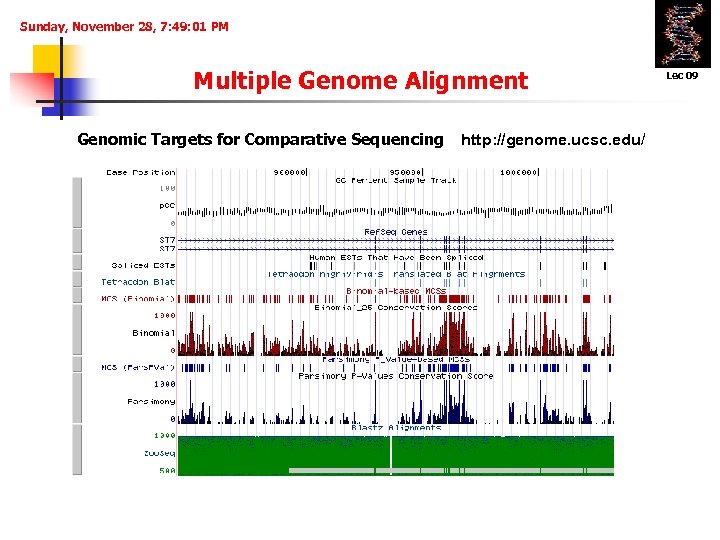 Sunday, November 28, 7: 49: 01 PM Multiple Genome Alignment Genomic Targets for Comparative Sequencing http: //genome. ucsc. edu/ Lec 09
Sunday, November 28, 7: 49: 01 PM Multiple Genome Alignment Genomic Targets for Comparative Sequencing http: //genome. ucsc. edu/ Lec 09


