5c02e4138e1ecc48d77c16d72781ff07.ppt
- Количество слайдов: 26

VIRTUAL LABORATORY AND ITS APPLICATION IN GENOMICS Luiza Handschuh Karol Marcinkowski University of Medical Sciences, Department of Haematology Institute of Bioorganic Chemistry PAS, Center of Excellence for Nucleic Acid-based Technologies INGRID 2008, Lacco Ameno, 11. 04. 2008

Virtual Laboratory - definition and advanteges „The Virtual Laboratory is a distributed workgroup environment, with the main task of providing a remote access to the various kind of rare and expensive scientific laboratory equipment and computational resources” (http: //vlab. psnc. pl/) q A specific representative of the RIS (Remote Instrumentation Systems) q Based on grid environment, already implemented in the VLab System by Poznan Supercomputing and Networking Center (PSNC) q Independent on physical location of the instruments q Designed to cooperate with many other grid systems q Devoted to experimental and computational tasks – supporting the postprocessing phase of experiment q Experiments made in other laboratories and their results can be shared enabling the workgroup
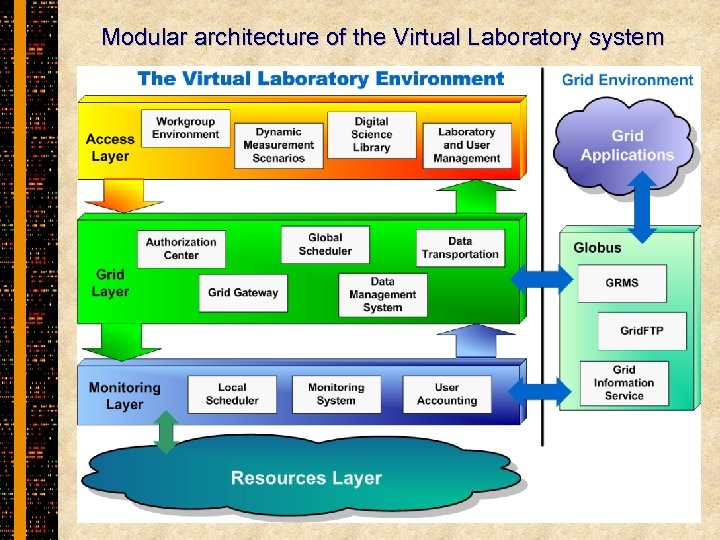
Modular architecture of the Virtual Laboratory system
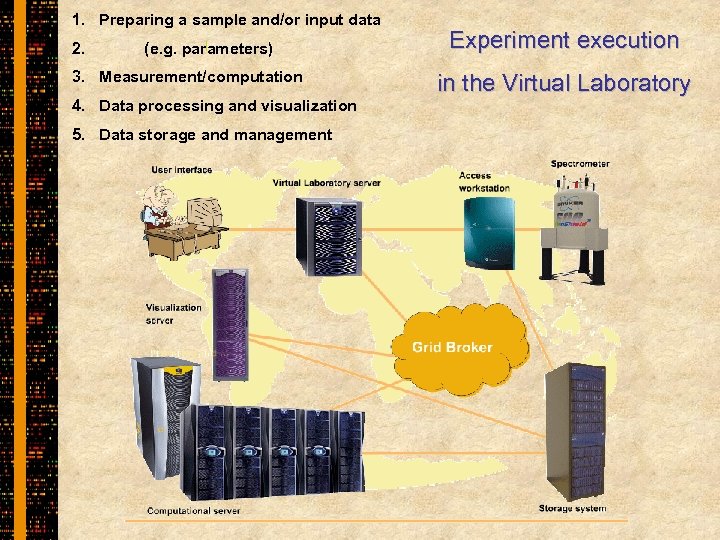
1. Preparing a sample and/or input data 2. (e. g. parameters) 3. Measurement/computation 4. Data processing and visualization 5. Data storage and management Experiment execution in the Virtual Laboratory
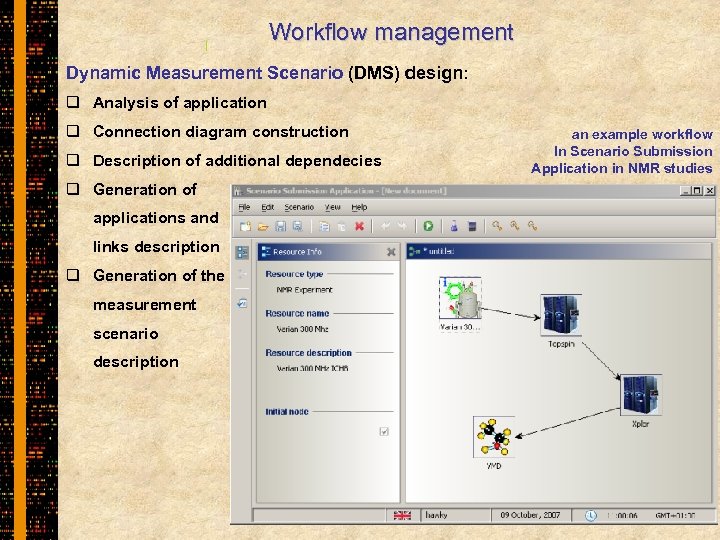
Workflow management Dynamic Measurement Scenario (DMS) design: q Analysis of application q Connection diagram construction q Description of additional dependecies q Generation of applications and links description q Generation of the measurement scenario description an example workflow In Scenario Submission Application in NMR studies
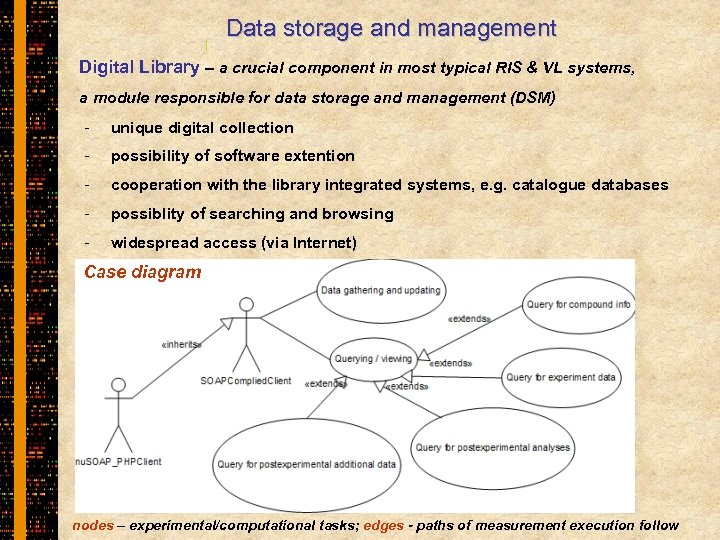
Data storage and management Digital Library – a crucial component in most typical RIS & VL systems, a module responsible for data storage and management (DSM) - unique digital collection - possibility of software extention - cooperation with the library integrated systems, e. g. catalogue databases - possiblity of searching and browsing - widespread access (via Internet) Case diagram nodes – experimental/computational tasks; edges - paths of measurement execution follow
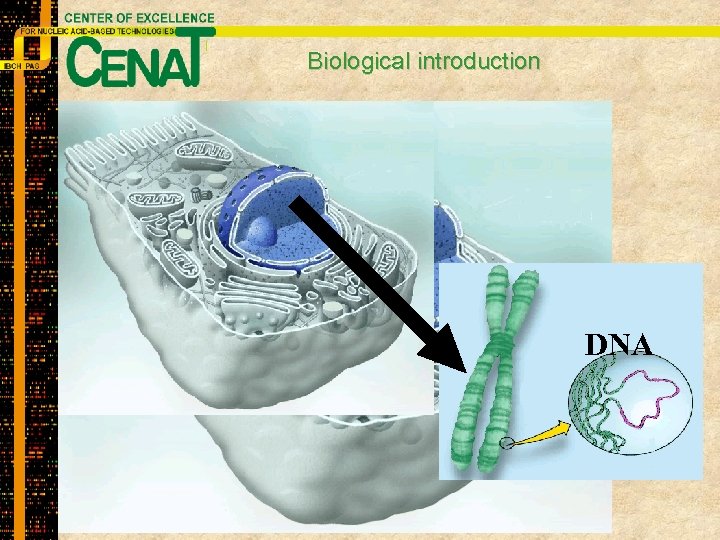
Biological introduction Mitochondria Cell nucleus Endoplasmatic reticulum Golgi Apparatus Tissues Organism Organs Plasma membrane DNA
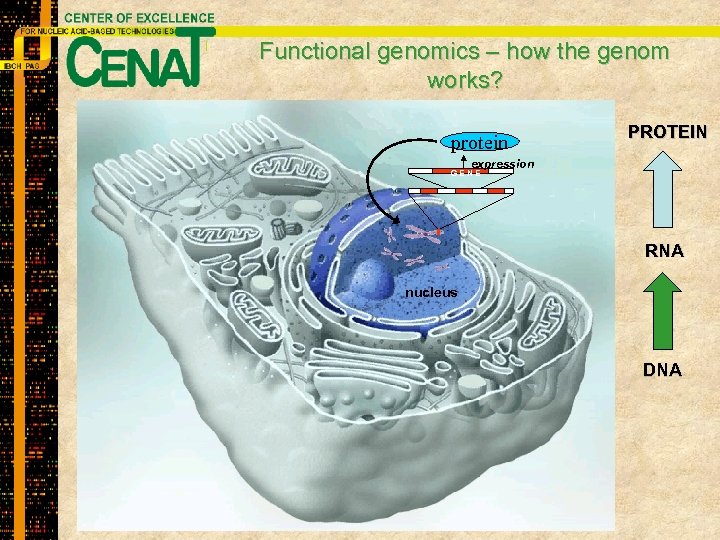
Functional genomics – how the genom works? protein PROTEIN expression GENE RNA nucleus DNA
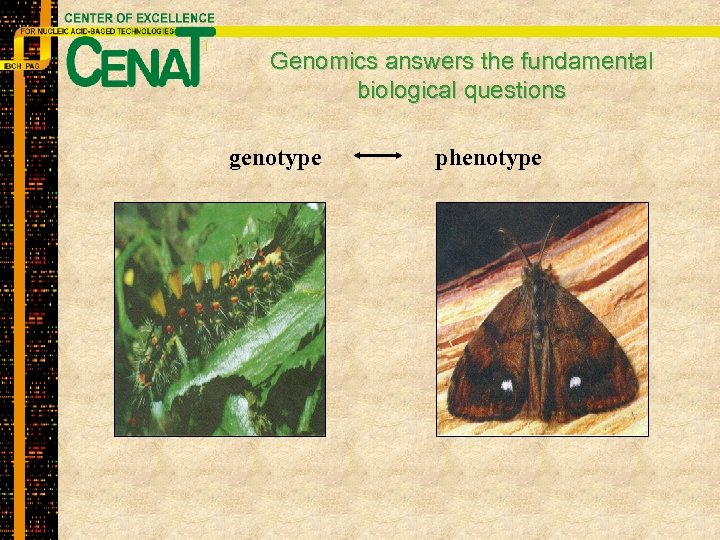
Genomics answers the fundamental biological questions genotype phenotype
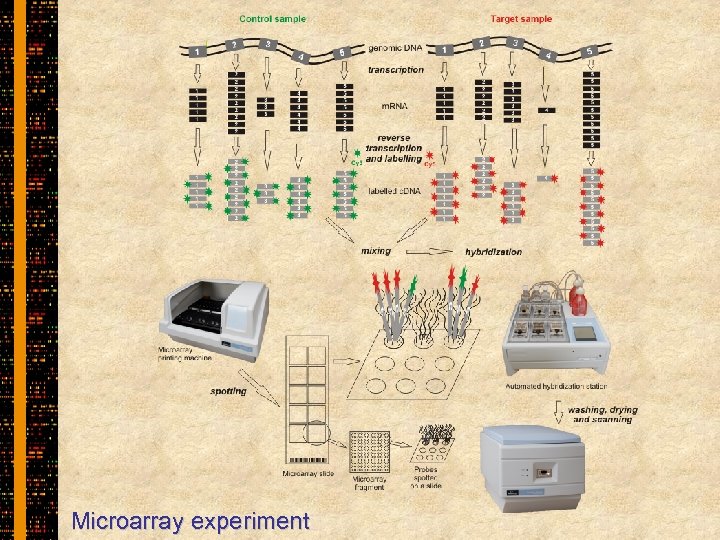
Microarray experiment
„Application of functional genomics tools for establishing complex model of tumor transformation. Studies on molecular mechanisms of acute myeloid leukemia pathogenesis” as a part of a huge project announced by Polish Ministry of Science and Informatization in 2005: „Application of functional genomics and bioinformatics for creation and characterisation of models describing biological processes of great importance in medicine and agriculture” (PBZ-MNi. I-2/1/2005) Institute of Bioorganic Chemistry PAS, Poznań Karol Marcinkowski University of Medical Sciences, Department of Haematology Poznań Supercomputing and Networking Center Poznań University of Technology
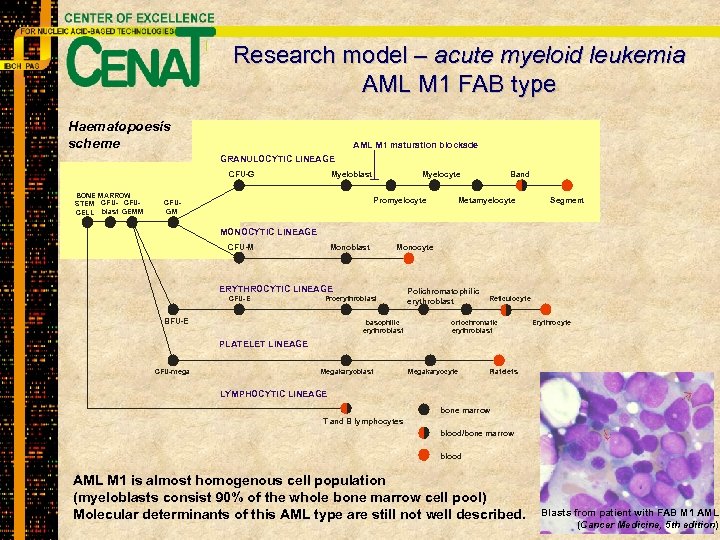
Research model – acute myeloid leukemia AML M 1 FAB type Haematopoesis scheme AML M 1 maturation blockade GRANULOCYTIC LINEAGE CFU-G BONE MARROW STEM CFU- CFUCELL blast GEMM Myeloblast Myelocyte Promyelocyte CFUGM Band Metamyelocyte Segment MONOCYTIC LINEAGE CFU-M Monoblast Monocyte ERYTHROCYTIC LINEAGE CFU-E Proerythroblast BFU-E basophilic erythroblast Polichromatophilic erythroblast Reticulocyte ortochromatic erythroblast Erythrocyte PLATELET LINEAGE CFU-mega Megakaryoblast Megakaryocyte Platelets LYMPHOCYTIC LINEAGE bone marrow T and B lymphocytes blood/bone marrow blood AML M 1 is almost homogenous cell population (myeloblasts consist 90% of the whole bone marrow cell pool) Molecular determinants of this AML type are still not well described. Blasts from patient with FAB M 1 AML (Cancer Medicine, 5 th edition)
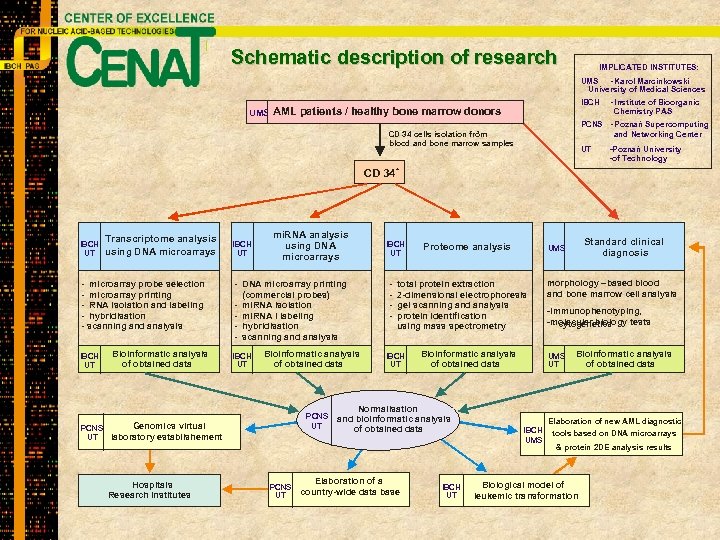
Schematic description of research IMPLICATED INSTITUTES: UMS - Karol Marcinkowski University of Medical Sciences UMS IBCH AML patients / healthy bone marrow donors PCNS - Poznań Supercomputing and Networking Center + CD 34 cells isolation from blood and bone marrow samples CD 34 IBCH UT Transcriptome analysis using DNA microarrays - microarray probe selection - microarray printing - RNA isolation and labeling - hybridisation - scanning and analysis IBCH UT Bioinformatic analysis of obtained data IBCH UT mi. RNA analysis using DNA microarrays - DNA microarray printing (commercial probes) - mi. RNA isolation - mi. RNA i labeling - hybridisation - scanning and analysis IBCH UT Bioinformatic analysis of obtained data PCNS UT Genomics virtual PCNS laboratory establishement UT Hospitals Research institutes PCNS UT UT -Poznań University -of Technology + IBCH UT - - Institute of Bioorganic Chemistry PAS Proteome analysis total protein extraction 2 -dimensional electrophoresis gel scanning and analysis protein identification using mass spectrometry IBCH UT Bioinformatic analysis of obtained data Normalisation and bioinformatic analysis of obtained data Elaboration of a country-wide data base IBCH UT Standard clinical diagnosis UMS morphology –based blood and bone marrow cell analysis -immunophenotyping, -molecular biology tests - cytogenetics UMS UT Bioinformatic analysis of obtained data Elaboration of new AML diagnostic IBCH UMS tools based on DNA microarrays & protein 2 DE analysis results Biological model of leukemic transformation
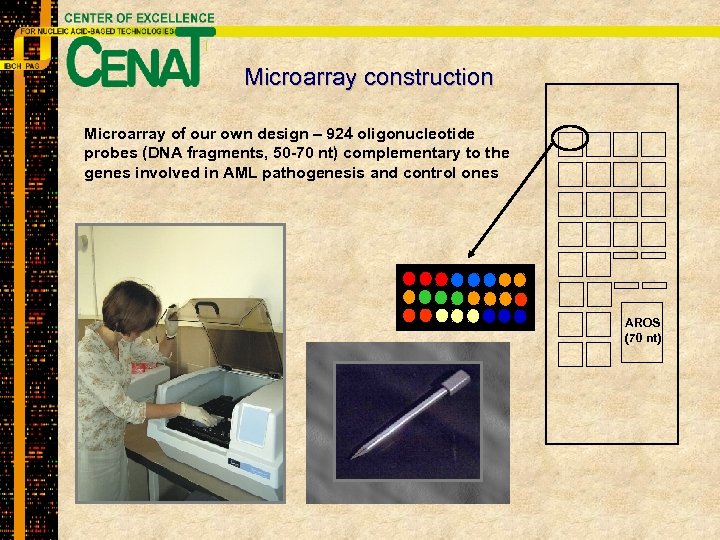
Microarray construction Microarray of our own design – 924 oligonucleotide probes (DNA fragments, 50 -70 nt) complementary to the genes involved in AML pathogenesis and control ones AROS (70 nt)
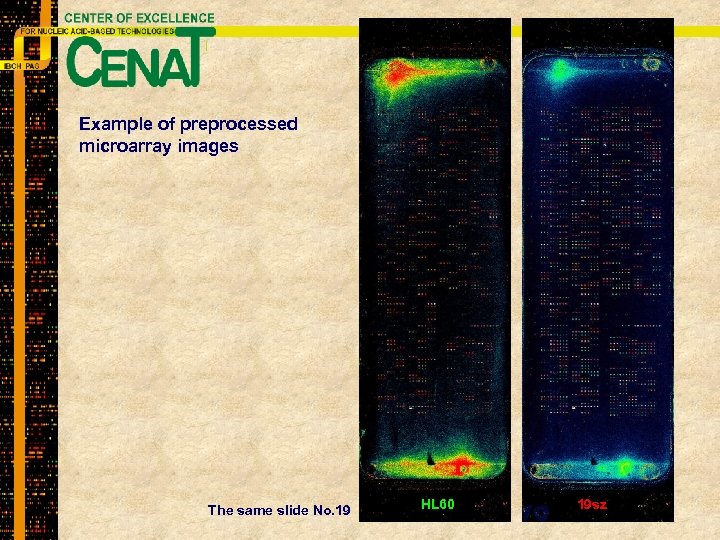
Example of preprocessed microarray images The same slide No. 19 HL 60 19 sz
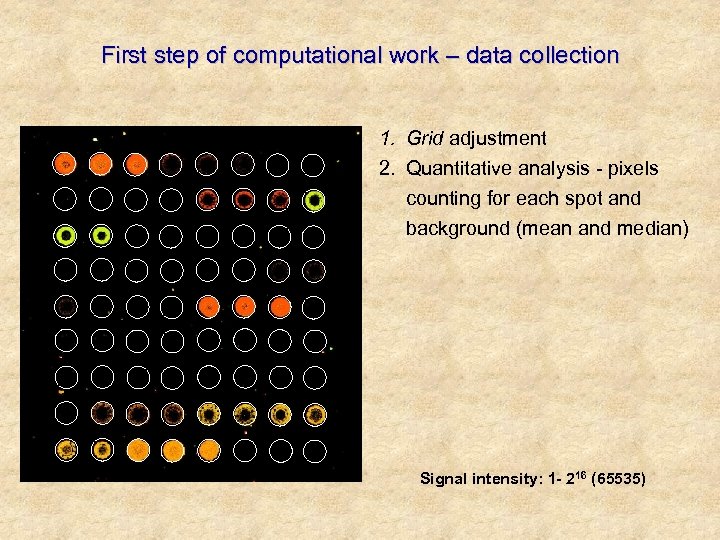
First step of computational work – data collection 1. Grid adjustment 2. Quantitative analysis - pixels counting for each spot and background (mean and median) Signal intensity: 1 - 216 (65535)
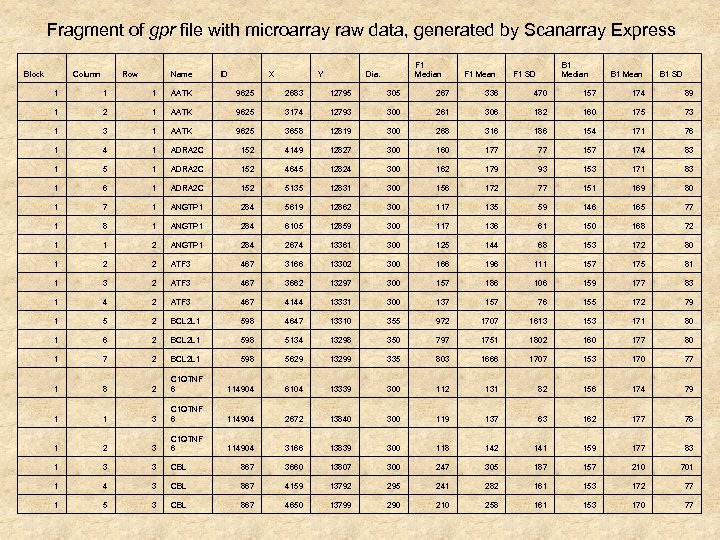
Fragment of gpr file with microarray raw data, generated by Scanarray Express Block Column Row Name ID X Y F 1 Median Dia. F 1 Mean B 1 Median F 1 SD B 1 Mean B 1 SD 1 1 1 AATK 9625 2683 12795 305 267 336 470 157 174 89 1 2 1 AATK 9625 3174 12793 300 261 306 182 160 175 73 1 AATK 9625 3658 12819 300 268 316 186 154 171 76 1 4 1 ADRA 2 C 152 4149 12827 300 160 177 77 157 174 83 1 5 1 ADRA 2 C 152 4645 12824 300 162 179 93 153 171 83 1 6 1 ADRA 2 C 152 5135 12831 300 156 172 77 151 169 80 1 7 1 ANGTP 1 284 5619 12862 300 117 135 59 146 165 77 1 8 1 ANGTP 1 284 6105 12859 300 117 136 61 150 168 72 1 1 2 ANGTP 1 284 2674 13361 300 125 144 68 153 172 80 1 2 2 ATF 3 467 3166 13302 300 166 196 111 157 175 81 1 3 2 ATF 3 467 3662 13297 300 157 186 106 159 177 83 1 4 2 ATF 3 467 4144 13331 300 137 157 76 155 172 79 1 5 2 BCL 2 L 1 598 4647 13310 355 972 1707 1613 153 171 80 1 6 2 BCL 2 L 1 598 5134 13298 350 797 1751 1802 160 177 80 1 7 2 BCL 2 L 1 598 5629 13299 335 803 1666 1707 153 170 77 1 8 2 C 1 QTNF 6 114904 6104 13339 300 112 131 82 156 174 79 1 1 3 C 1 QTNF 6 114904 2672 13840 300 119 137 63 162 177 78 1 2 3 C 1 QTNF 6 114904 3166 13839 300 118 142 141 159 177 83 1 3 3 CBL 867 3660 13807 300 247 305 187 157 210 701 1 4 3 CBL 867 4159 13792 295 241 282 161 153 172 77 1 5 3 CBL 867 4650 13799 290 210 258 161 153 170 77
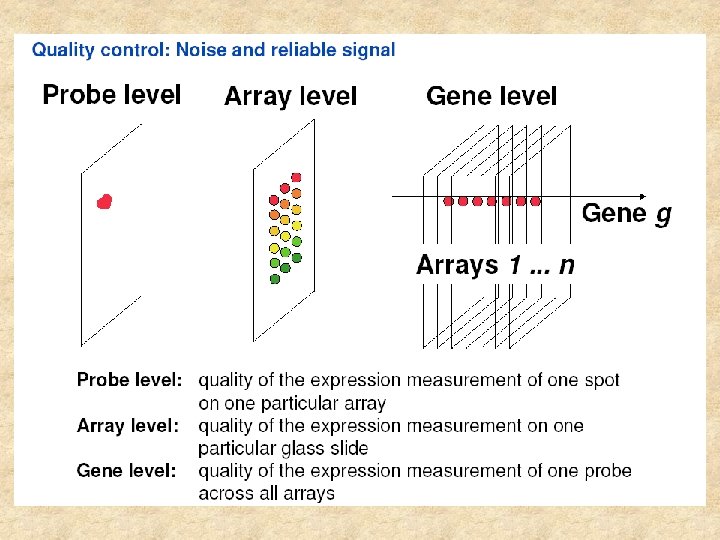
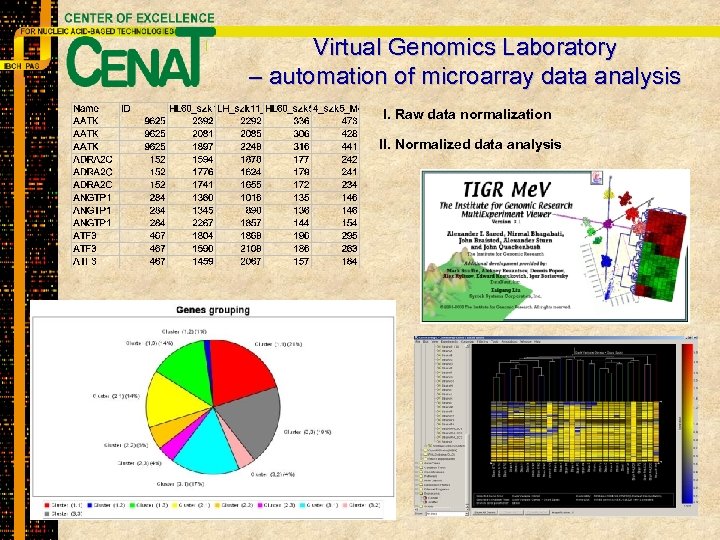
Virtual Genomics Laboratory – automation of microarray data analysis I. Raw data normalization II. Normalized data analysis
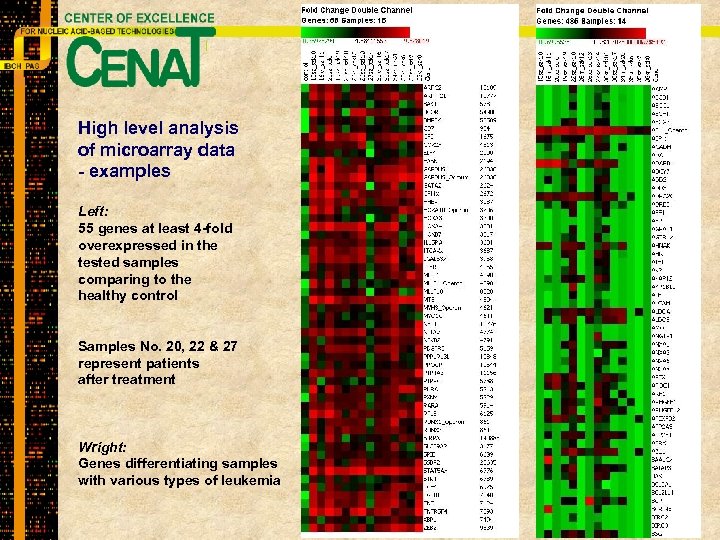
High level analysis of microarray data - examples Left: 55 genes at least 4 -fold overexpressed in the tested samples comparing to the healthy control Samples No. 20, 22 & 27 represent patients after treatment Wright: Genes differentiating samples with various types of leukemia
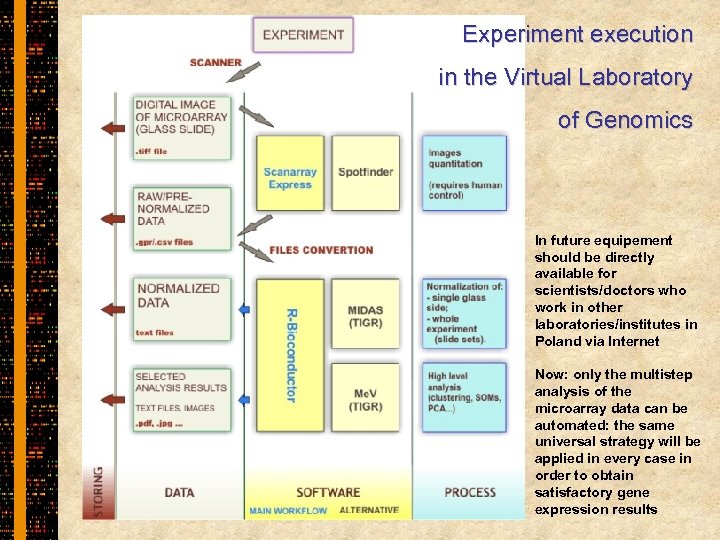
Experiment execution in the Virtual Laboratory of Genomics In future equipement should be directly available for scientists/doctors who work in other laboratories/institutes in Poland via Internet Now: only the multistep analysis of the microarray data can be automated: the same universal strategy will be applied in every case in order to obtain satisfactory gene expression results
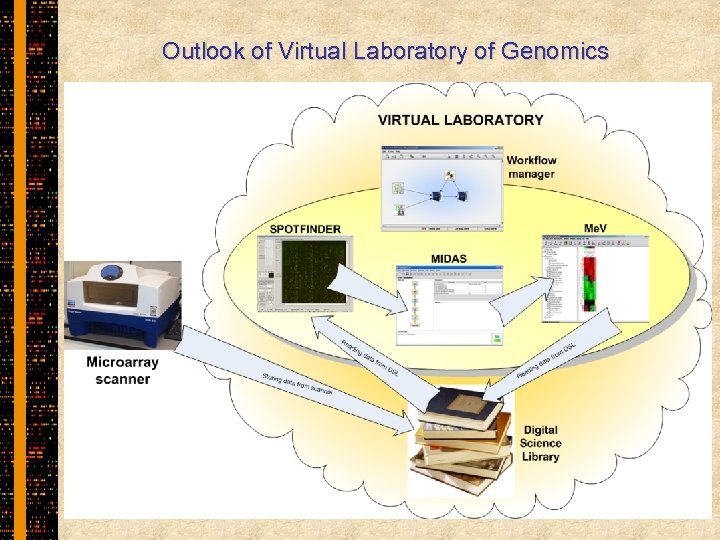
Outlook of Virtual Laboratory of Genomics

The authors of publication Marcin Lawenda, Norbert Meyer, Maciej Stroinski, Jan Weglarz Poznan Supercomputing and Networking Center Luiza Handschuh, Piotr Stepniak, Marek Figlerowicz (director of the project) Institute of Bioorganic Chemistry PAS Others participants of the project: Maciej Kaźmierczak, Mieczysław Komarnicki, Krzysztof Lewandowski Karol Marcinkowski University of Medical Sciences, Departament of Haematology Piotr Formanowicz, Jacek Błazewicz Poznan University of Technology, Institute of Informatics

5c02e4138e1ecc48d77c16d72781ff07.ppt