35358ae9f3030606d0e687df98bcf07e.ppt
- Количество слайдов: 16
 Using Chem. Axon Toolkits in the Lead Discovery Database at GNF Hayk Asatryan, Dimitri Petrov, S. Frank Yan, Andrey Santrosyan, Kaisheng Chen, Shumei Jiang, Jeff Janes, Yingyao Zhou May, 2005 Budapest, Hungry Genomics Institute of the Novartis Research Foundation
Using Chem. Axon Toolkits in the Lead Discovery Database at GNF Hayk Asatryan, Dimitri Petrov, S. Frank Yan, Andrey Santrosyan, Kaisheng Chen, Shumei Jiang, Jeff Janes, Yingyao Zhou May, 2005 Budapest, Hungry Genomics Institute of the Novartis Research Foundation
 Scope of the Lead Discovery Database - LDDB Compound Management Center HTS Center Data Processing Quality Control Hit Picking Program Management Hit to Lead Tracking Analytical Chemistry Med. Chem QSAR Optimized Leads ADME/Tox PK/PD Genomics Institute of the Novartis Research Foundation
Scope of the Lead Discovery Database - LDDB Compound Management Center HTS Center Data Processing Quality Control Hit Picking Program Management Hit to Lead Tracking Analytical Chemistry Med. Chem QSAR Optimized Leads ADME/Tox PK/PD Genomics Institute of the Novartis Research Foundation
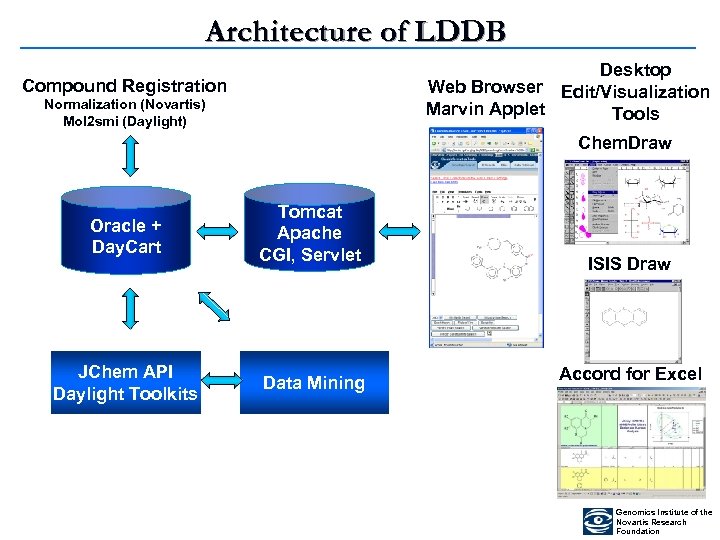 Architecture of LDDB Desktop Web Browser Edit/Visualization Marvin Applet Tools Compound Registration Normalization (Novartis) Mol 2 smi (Daylight) Chem. Draw Oracle + Day. Cart Tomcat Apache CGI, Servlet JChem API Daylight Toolkits Data Mining ISIS Draw Accord for Excel Genomics Institute of the Novartis Research Foundation
Architecture of LDDB Desktop Web Browser Edit/Visualization Marvin Applet Tools Compound Registration Normalization (Novartis) Mol 2 smi (Daylight) Chem. Draw Oracle + Day. Cart Tomcat Apache CGI, Servlet JChem API Daylight Toolkits Data Mining ISIS Draw Accord for Excel Genomics Institute of the Novartis Research Foundation
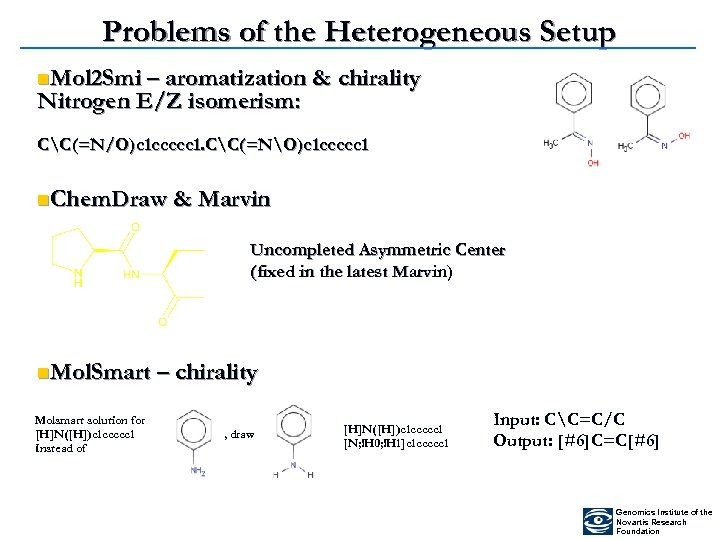 Problems of the Heterogeneous Setup n. Mol 2 Smi – aromatization & Nitrogen E/Z isomerism: chirality CC(=N/O)c 1 ccccc 1. CC(=NO)c 1 ccccc 1 n. Chem. Draw & Marvin Uncompleted Asymmetric Center (fixed in the latest Marvin) n. Mol. Smart – chirality Molsmart solution for [H]N([H])c 1 ccccc 1 Instead of , draw [H]N([H])c 1 ccccc 1 [N; !H 0; !H 1]c 1 ccccc 1 Input: CC=C/C Output: [#6]C=C[#6] Genomics Institute of the Novartis Research Foundation
Problems of the Heterogeneous Setup n. Mol 2 Smi – aromatization & Nitrogen E/Z isomerism: chirality CC(=N/O)c 1 ccccc 1. CC(=NO)c 1 ccccc 1 n. Chem. Draw & Marvin Uncompleted Asymmetric Center (fixed in the latest Marvin) n. Mol. Smart – chirality Molsmart solution for [H]N([H])c 1 ccccc 1 Instead of , draw [H]N([H])c 1 ccccc 1 [N; !H 0; !H 1]c 1 ccccc 1 Input: CC=C/C Output: [#6]C=C[#6] Genomics Institute of the Novartis Research Foundation
 Problems of the Heterogeneous Setup (cont. ) n. Daylight & Chem. Axon (discuss later) n. Accord: chirality, display Chem. Draw Accord for Excel Marvin n. Pricing considerations Genomics Institute of the Novartis Research Foundation
Problems of the Heterogeneous Setup (cont. ) n. Daylight & Chem. Axon (discuss later) n. Accord: chirality, display Chem. Draw Accord for Excel Marvin n. Pricing considerations Genomics Institute of the Novartis Research Foundation
 JChem Cartridge – initial testing (July 2004) Daylight's substructure search: 5 -6 seconds JCart substructure search: 10 -12 sec (caching the whole structure table in Oracle) Similarity search is approximately 40 -50 minutes (1. 76 million) JChem results: 10. 6 minutes for 3 million structures (3 GHz Pentium 4) Smiles NC(=O)C(=NOC)c 1 csc(N)n 1 Count Time (ms) 52 12044 OC(=O)c 1 cc(O)c(O)c 1 530 12091 OC 1 CC(C)(N)C(O)C(C)O 1 3 11420 48 10873 0 10310 36 11482 283 14216 2 10857 NC(=O)C(N)Cc 1 cnc[n. H]1 315 18011 OC(=O)c 1 cc(OC)c(OC)c 1 436 11779 OC 1 OC(CO)C(OC(N)=O)C 1 O 2 10764 c 1 c(C)[n. H]c(=O)[n. H]c 1=O 0 10919 c 1 ccc(OS(=O)O)cc 1 C 2 OC(n 1 ccc(=O)[n. H]c 1=O)C(O)C 2 O Cc 1 ccc(N(CCCl)cc 1 OC 1 OC(C)C(OC)C 1 OC(CO)C(OC(N)=O)C 1 O C(=O)NC(C=O)CCCNC(=N)N 1835 Genomics Institute 15184 Research of the Novartis Foundation
JChem Cartridge – initial testing (July 2004) Daylight's substructure search: 5 -6 seconds JCart substructure search: 10 -12 sec (caching the whole structure table in Oracle) Similarity search is approximately 40 -50 minutes (1. 76 million) JChem results: 10. 6 minutes for 3 million structures (3 GHz Pentium 4) Smiles NC(=O)C(=NOC)c 1 csc(N)n 1 Count Time (ms) 52 12044 OC(=O)c 1 cc(O)c(O)c 1 530 12091 OC 1 CC(C)(N)C(O)C(C)O 1 3 11420 48 10873 0 10310 36 11482 283 14216 2 10857 NC(=O)C(N)Cc 1 cnc[n. H]1 315 18011 OC(=O)c 1 cc(OC)c(OC)c 1 436 11779 OC 1 OC(CO)C(OC(N)=O)C 1 O 2 10764 c 1 c(C)[n. H]c(=O)[n. H]c 1=O 0 10919 c 1 ccc(OS(=O)O)cc 1 C 2 OC(n 1 ccc(=O)[n. H]c 1=O)C(O)C 2 O Cc 1 ccc(N(CCCl)cc 1 OC 1 OC(C)C(OC)C 1 OC(CO)C(OC(N)=O)C 1 O C(=O)NC(C=O)CCCNC(=N)N 1835 Genomics Institute 15184 Research of the Novartis Foundation
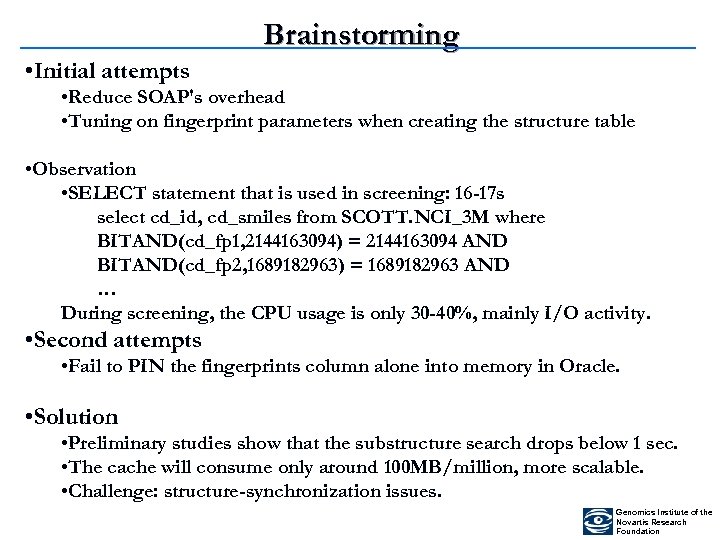 Brainstorming • Initial attempts • Reduce SOAP's overhead • Tuning on fingerprint parameters when creating the structure table • Observation • SELECT statement that is used in screening: 16 -17 s select cd_id, cd_smiles from SCOTT. NCI_3 M where BITAND(cd_fp 1, 2144163094) = 2144163094 AND BITAND(cd_fp 2, 1689182963) = 1689182963 AND … During screening, the CPU usage is only 30 -40%, mainly I/O activity. • Second attempts • Fail to PIN the fingerprints column alone into memory in Oracle. • Solution • Preliminary studies show that the substructure search drops below 1 sec. • The cache will consume only around 100 MB/million, more scalable. • Challenge: structure-synchronization issues. Genomics Institute of the Novartis Research Foundation
Brainstorming • Initial attempts • Reduce SOAP's overhead • Tuning on fingerprint parameters when creating the structure table • Observation • SELECT statement that is used in screening: 16 -17 s select cd_id, cd_smiles from SCOTT. NCI_3 M where BITAND(cd_fp 1, 2144163094) = 2144163094 AND BITAND(cd_fp 2, 1689182963) = 1689182963 AND … During screening, the CPU usage is only 30 -40%, mainly I/O activity. • Second attempts • Fail to PIN the fingerprints column alone into memory in Oracle. • Solution • Preliminary studies show that the substructure search drops below 1 sec. • The cache will consume only around 100 MB/million, more scalable. • Challenge: structure-synchronization issues. Genomics Institute of the Novartis Research Foundation
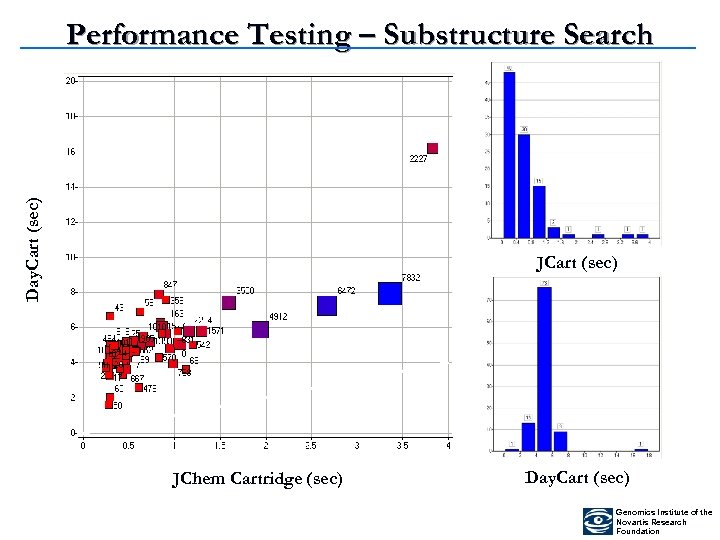 Day. Cart (sec) Performance Testing – Substructure Search JCart (sec) JChem Cartridge (sec) Day. Cart (sec) Genomics Institute of the Novartis Research Foundation
Day. Cart (sec) Performance Testing – Substructure Search JCart (sec) JChem Cartridge (sec) Day. Cart (sec) Genomics Institute of the Novartis Research Foundation
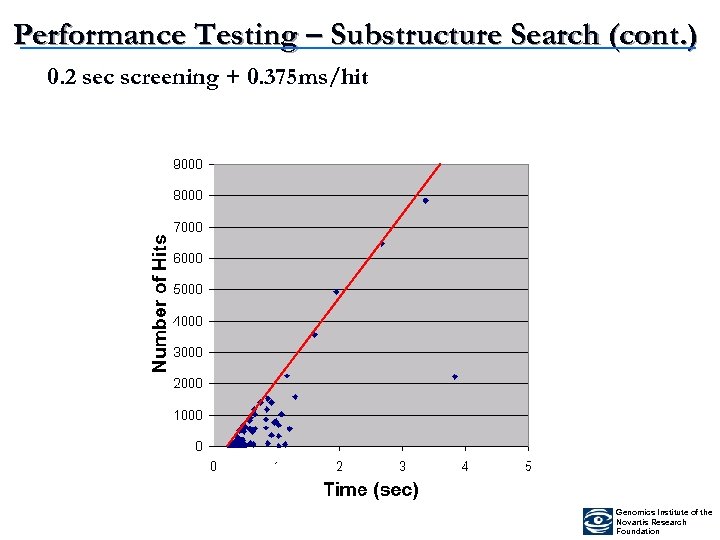 Performance Testing – Substructure Search (cont. ) 0. 2 sec screening + 0. 375 ms/hit Genomics Institute of the Novartis Research Foundation
Performance Testing – Substructure Search (cont. ) 0. 2 sec screening + 0. 375 ms/hit Genomics Institute of the Novartis Research Foundation
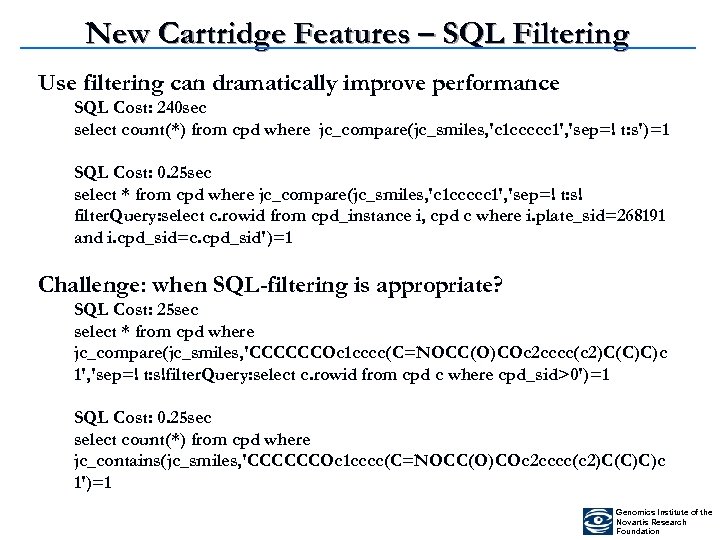 New Cartridge Features – SQL Filtering Use filtering can dramatically improve performance SQL Cost: 240 sec select count(*) from cpd where jc_compare(jc_smiles, 'c 1 ccccc 1', 'sep=! t: s')=1 SQL Cost: 0. 25 sec select * from cpd where jc_compare(jc_smiles, 'c 1 ccccc 1', 'sep=! t: s! filter. Query: select c. rowid from cpd_instance i, cpd c where i. plate_sid=268191 and i. cpd_sid=c. cpd_sid')=1 Challenge: when SQL-filtering is appropriate? SQL Cost: 25 sec select * from cpd where jc_compare(jc_smiles, 'CCCCCCOc 1 cccc(C=NOCC(O)COc 2 cccc(c 2)C(C)C)c 1', 'sep=! t: s!filter. Query: select c. rowid from cpd c where cpd_sid>0')=1 SQL Cost: 0. 25 sec select count(*) from cpd where jc_contains(jc_smiles, 'CCCCCCOc 1 cccc(C=NOCC(O)COc 2 cccc(c 2)C(C)C)c 1')=1 Genomics Institute of the Novartis Research Foundation
New Cartridge Features – SQL Filtering Use filtering can dramatically improve performance SQL Cost: 240 sec select count(*) from cpd where jc_compare(jc_smiles, 'c 1 ccccc 1', 'sep=! t: s')=1 SQL Cost: 0. 25 sec select * from cpd where jc_compare(jc_smiles, 'c 1 ccccc 1', 'sep=! t: s! filter. Query: select c. rowid from cpd_instance i, cpd c where i. plate_sid=268191 and i. cpd_sid=c. cpd_sid')=1 Challenge: when SQL-filtering is appropriate? SQL Cost: 25 sec select * from cpd where jc_compare(jc_smiles, 'CCCCCCOc 1 cccc(C=NOCC(O)COc 2 cccc(c 2)C(C)C)c 1', 'sep=! t: s!filter. Query: select c. rowid from cpd c where cpd_sid>0')=1 SQL Cost: 0. 25 sec select count(*) from cpd where jc_contains(jc_smiles, 'CCCCCCOc 1 cccc(C=NOCC(O)COc 2 cccc(c 2)C(C)C)c 1')=1 Genomics Institute of the Novartis Research Foundation
 Day. Cart to JCart Migration Challenges n n Identical structures or not? Two identical structures considered by Daylight ideally remains identical by JChem, and vice versa. Example 1: Aromatic Sulfur Solution: Jchem support Daylight rules COC 1=NC(=NS(=N 1)C 2 CCCCC 2)Cl. C Oc 1 nc(Cl)ns(n 1)C 2 CCCCC 2 Genomics Institute of the Novartis Research Foundation
Day. Cart to JCart Migration Challenges n n Identical structures or not? Two identical structures considered by Daylight ideally remains identical by JChem, and vice versa. Example 1: Aromatic Sulfur Solution: Jchem support Daylight rules COC 1=NC(=NS(=N 1)C 2 CCCCC 2)Cl. C Oc 1 nc(Cl)ns(n 1)C 2 CCCCC 2 Genomics Institute of the Novartis Research Foundation
![Challenges – Identical Structures? (cont. ) n. Example 2: Isotope Bug [2 H][C@H]1 O[C@H]1 Challenges – Identical Structures? (cont. ) n. Example 2: Isotope Bug [2 H][C@H]1 O[C@H]1](https://present5.com/presentation/35358ae9f3030606d0e687df98bcf07e/image-12.jpg) Challenges – Identical Structures? (cont. ) n. Example 2: Isotope Bug [2 H][C@H]1 O[C@H]1 COCc 2 ccccc 2 C(OCc 1 ccccc 1)[C@H]2 CO 2 n. Example 3: Standardization CC 1=CC=N(C)C=C 1 Cc 1 cc[n+](C)cc 1 Genomics Institute of the Novartis Research Foundation
Challenges – Identical Structures? (cont. ) n. Example 2: Isotope Bug [2 H][C@H]1 O[C@H]1 COCc 2 ccccc 2 C(OCc 1 ccccc 1)[C@H]2 CO 2 n. Example 3: Standardization CC 1=CC=N(C)C=C 1 Cc 1 cc[n+](C)cc 1 Genomics Institute of the Novartis Research Foundation
 Challenges – Identical Structures? (cont. ) n. Example 4: Non-standard Bond Brc 1 ccc 2[N]c 3 nc 4 ccccc 4 nc 3 -c 2 c 1 Brc 1 ccc 2 Nc 3 nc 4 ccccc 4 nc 3 -c 2 c 1 n. Example 4: Chirality C[C@]1(O)C[C@@]23 C[C@H](O)C 4 C(CCC 5=CC(=O)CC[C@]45 C)C 3 CCC 12 n. Incomplete Structures in Database *c 1 ccc(COCC 2 CCC=CO 2)cc 1 or NULL (not supported by JCart) Genomics Institute of the Novartis Research Foundation
Challenges – Identical Structures? (cont. ) n. Example 4: Non-standard Bond Brc 1 ccc 2[N]c 3 nc 4 ccccc 4 nc 3 -c 2 c 1 Brc 1 ccc 2 Nc 3 nc 4 ccccc 4 nc 3 -c 2 c 1 n. Example 4: Chirality C[C@]1(O)C[C@@]23 C[C@H](O)C 4 C(CCC 5=CC(=O)CC[C@]45 C)C 3 CCC 12 n. Incomplete Structures in Database *c 1 ccc(COCC 2 CCC=CO 2)cc 1 or NULL (not supported by JCart) Genomics Institute of the Novartis Research Foundation
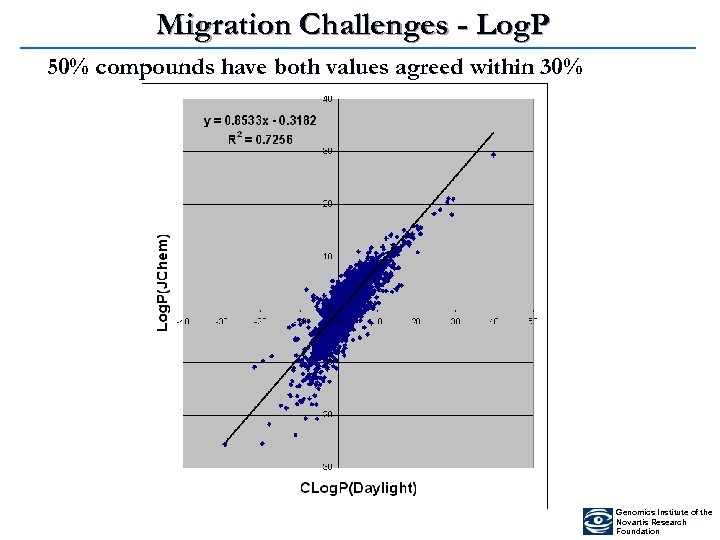 Migration Challenges - Log. P 50% compounds have both values agreed within 30% Genomics Institute of the Novartis Research Foundation
Migration Challenges - Log. P 50% compounds have both values agreed within 30% Genomics Institute of the Novartis Research Foundation
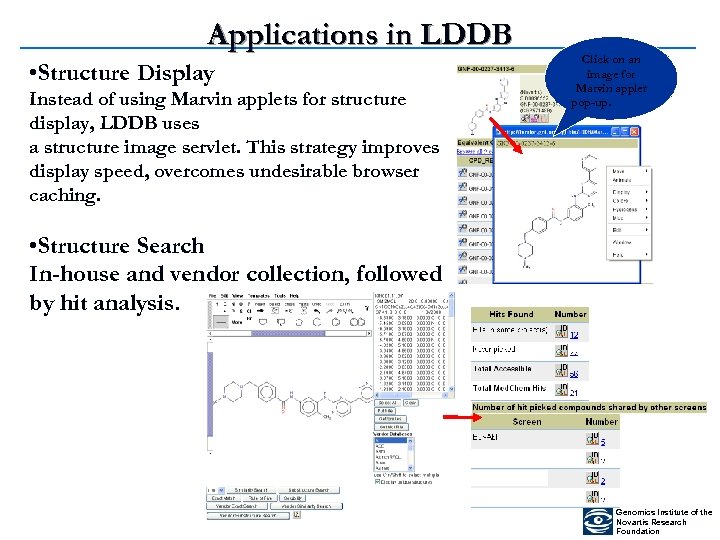 Applications in LDDB • Structure Display Instead of using Marvin applets for structure display, LDDB uses a structure image servlet. This strategy improves display speed, overcomes undesirable browser caching. Click on an image for Marvin applet pop-up. • Structure Search In-house and vendor collection, followed by hit analysis. Genomics Institute of the Novartis Research Foundation
Applications in LDDB • Structure Display Instead of using Marvin applets for structure display, LDDB uses a structure image servlet. This strategy improves display speed, overcomes undesirable browser caching. Click on an image for Marvin applet pop-up. • Structure Search In-house and vendor collection, followed by hit analysis. Genomics Institute of the Novartis Research Foundation
 Ongoing Developments … • Other applications R-group analysis Most-common substructure analysis Database-wise clustering analysis Thank You! ZHOU@GNF. ORG Genomics Institute of the Novartis Research Foundation
Ongoing Developments … • Other applications R-group analysis Most-common substructure analysis Database-wise clustering analysis Thank You! ZHOU@GNF. ORG Genomics Institute of the Novartis Research Foundation


