2ec06d03879e293ea0ab2f273f5fc88b.ppt
- Количество слайдов: 39
 Understanding Sequence, Structure and Function Relationships and the Resulting Redundancy PHAR 201/Bioinformatics I Philip E. Bourne UCSD PHAR 201 Lecture 07, 2012 1
Understanding Sequence, Structure and Function Relationships and the Resulting Redundancy PHAR 201/Bioinformatics I Philip E. Bourne UCSD PHAR 201 Lecture 07, 2012 1
 Agenda • Understand the relationship between sequence, structure and function. Consider specifically: – sequence-structure – structure-function • Take home message: a non-redundant set of sequences is different than a non-redundant set of structures is different than a non-redundant set of functions PHAR 201 Lecture 07, 2012 2
Agenda • Understand the relationship between sequence, structure and function. Consider specifically: – sequence-structure – structure-function • Take home message: a non-redundant set of sequences is different than a non-redundant set of structures is different than a non-redundant set of functions PHAR 201 Lecture 07, 2012 2
 Why Bother? • Biology: – A full understanding of a molecular system comes from careful examination of the sequence-structure-function triad – Each triad is then a component in a biological process • Method: – Bioinformatics studies invariably start from a non-redundant set of data to achieve appropriate statistical significance PHAR 201 Lecture 07, 2012 3
Why Bother? • Biology: – A full understanding of a molecular system comes from careful examination of the sequence-structure-function triad – Each triad is then a component in a biological process • Method: – Bioinformatics studies invariably start from a non-redundant set of data to achieve appropriate statistical significance PHAR 201 Lecture 07, 2012 3
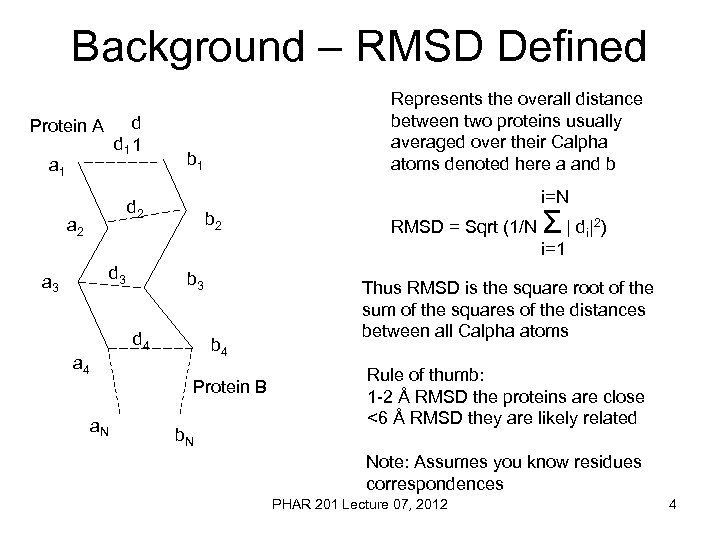 Background – RMSD Defined Protein A a 1 d d 1 1 b 1 i=N d 2 a 2 Represents the overall distance between two proteins usually averaged over their Calpha atoms denoted here a and b b 2 RMSD = Sqrt (1/N Σ | d| ) i=1 d 3 a 3 b 3 d 4 b 4 a 4 Protein B a. N b. N i 2 Thus RMSD is the square root of the sum of the squares of the distances between all Calpha atoms Rule of thumb: 1 -2 Å RMSD the proteins are close <6 Å RMSD they are likely related Note: Assumes you know residues correspondences PHAR 201 Lecture 07, 2012 4
Background – RMSD Defined Protein A a 1 d d 1 1 b 1 i=N d 2 a 2 Represents the overall distance between two proteins usually averaged over their Calpha atoms denoted here a and b b 2 RMSD = Sqrt (1/N Σ | d| ) i=1 d 3 a 3 b 3 d 4 b 4 a 4 Protein B a. N b. N i 2 Thus RMSD is the square root of the sum of the squares of the distances between all Calpha atoms Rule of thumb: 1 -2 Å RMSD the proteins are close <6 Å RMSD they are likely related Note: Assumes you know residues correspondences PHAR 201 Lecture 07, 2012 4
 Some Useful Observations • Below 30% protein sequence identity detection of a homologous relationship is not guaranteed by sequence alone • Structure is much more conserved than sequence • Distinguishing between divergent versus convergent evolution is an issue • Structure is limited relative to sequence or the order 1: 100 – 1: 10000 (depending on how you count) • Structure follows a power law with respect to function – each structural template has from 1 to n functions PHAR 201 Lecture 07, 2012 5
Some Useful Observations • Below 30% protein sequence identity detection of a homologous relationship is not guaranteed by sequence alone • Structure is much more conserved than sequence • Distinguishing between divergent versus convergent evolution is an issue • Structure is limited relative to sequence or the order 1: 100 – 1: 10000 (depending on how you count) • Structure follows a power law with respect to function – each structural template has from 1 to n functions PHAR 201 Lecture 07, 2012 5
 Relationship Between Sequence and Structure PHAR 201 Lecture 07, 2012 6
Relationship Between Sequence and Structure PHAR 201 Lecture 07, 2012 6
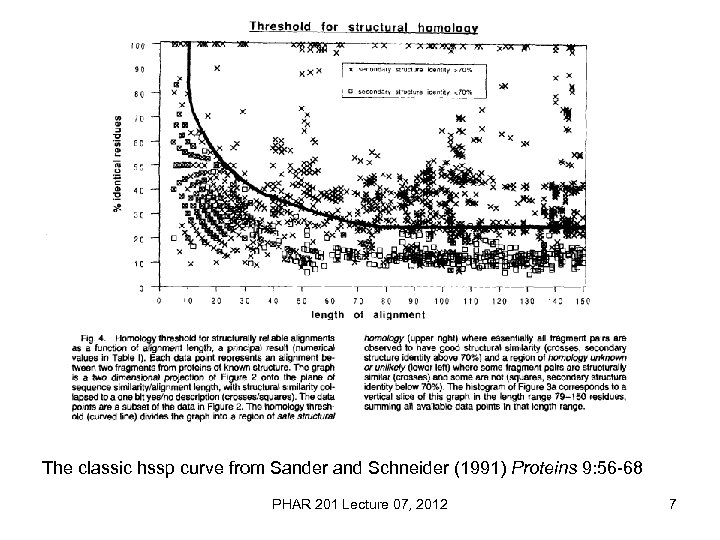 The classic hssp curve from Sander and Schneider (1991) Proteins 9: 56 -68 PHAR 201 Lecture 07, 2012 7
The classic hssp curve from Sander and Schneider (1991) Proteins 9: 56 -68 PHAR 201 Lecture 07, 2012 7
 This Analysis was Updated by Rost in 1999 http: //peds. oupjournals. org/cgi/con tent/full/12/2/85 PHAR 201 Lecture 07, 2012 8
This Analysis was Updated by Rost in 1999 http: //peds. oupjournals. org/cgi/con tent/full/12/2/85 PHAR 201 Lecture 07, 2012 8
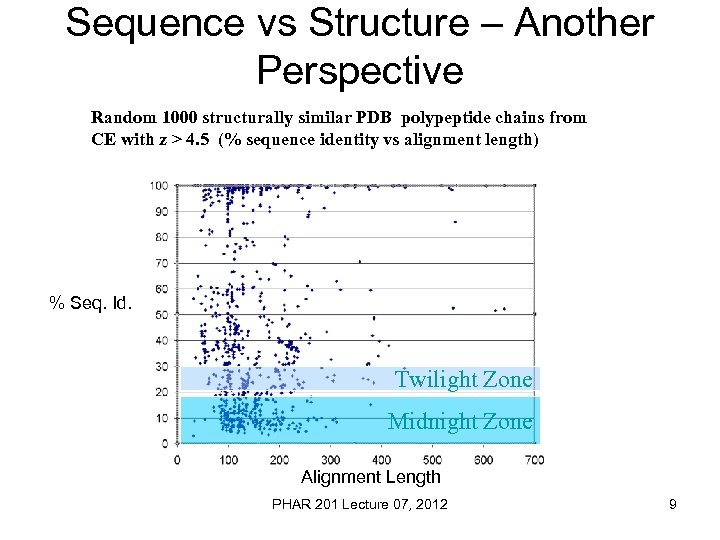 Sequence vs Structure – Another Perspective Random 1000 structurally similar PDB polypeptide chains from CE with z > 4. 5 (% sequence identity vs alignment length) % Seq. Id. Twilight Zone Midnight Zone Alignment Length PHAR 201 Lecture 07, 2012 9
Sequence vs Structure – Another Perspective Random 1000 structurally similar PDB polypeptide chains from CE with z > 4. 5 (% sequence identity vs alignment length) % Seq. Id. Twilight Zone Midnight Zone Alignment Length PHAR 201 Lecture 07, 2012 9
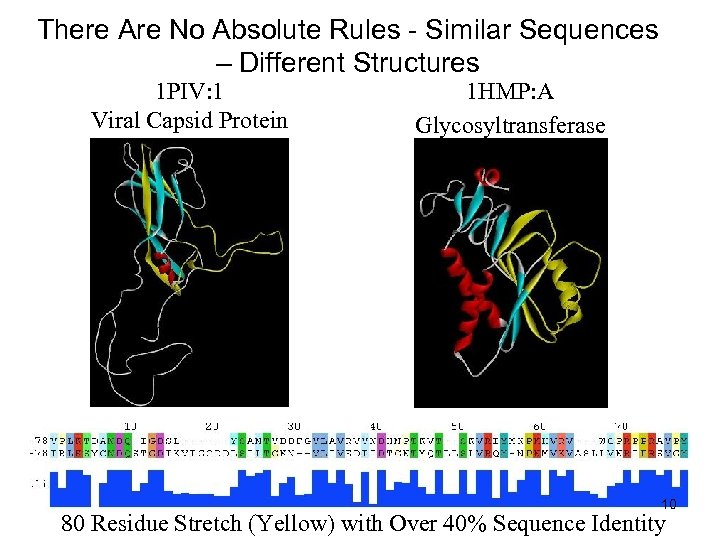 There Are No Absolute Rules - Similar Sequences – Different Structures 1 PIV: 1 Viral Capsid Protein 1 HMP: A Glycosyltransferase 10 80 Residue Stretch (Yellow) with Over 40% Sequence Identity
There Are No Absolute Rules - Similar Sequences – Different Structures 1 PIV: 1 Viral Capsid Protein 1 HMP: A Glycosyltransferase 10 80 Residue Stretch (Yellow) with Over 40% Sequence Identity
 Given This Complex Relationship a Non-redundant Set of Sequences Does not Imply a Non -redundant Set of Structures PHAR 201 Lecture 07, 2012 11
Given This Complex Relationship a Non-redundant Set of Sequences Does not Imply a Non -redundant Set of Structures PHAR 201 Lecture 07, 2012 11
 Structure vs Structure PHAR 201 Lecture 07, 2012 12
Structure vs Structure PHAR 201 Lecture 07, 2012 12
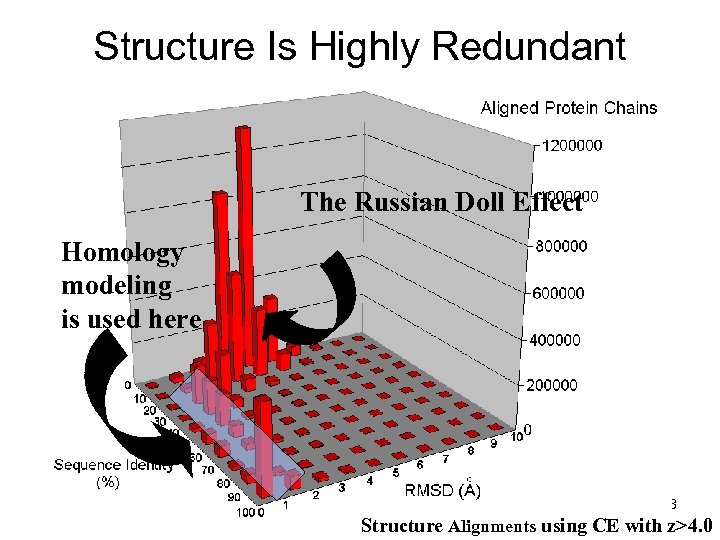 Structure Is Highly Redundant The Russian Doll Effect Homology modeling is used here PHAR 201 Lecture 07, 2012 13 Structure Alignments using CE with z>4. 0
Structure Is Highly Redundant The Russian Doll Effect Homology modeling is used here PHAR 201 Lecture 07, 2012 13 Structure Alignments using CE with z>4. 0
 We will be revisiting this in the next couple of lectures • Specifically: – How do we capture this redundancy? – What systems are commonly used to express this redundancy and what do they bring to our understanding of biology? • For now consider what this means using the most popular structure classification scheme - SCOP PHAR 201 Lecture 07, 2012 14
We will be revisiting this in the next couple of lectures • Specifically: – How do we capture this redundancy? – What systems are commonly used to express this redundancy and what do they bring to our understanding of biology? • For now consider what this means using the most popular structure classification scheme - SCOP PHAR 201 Lecture 07, 2012 14
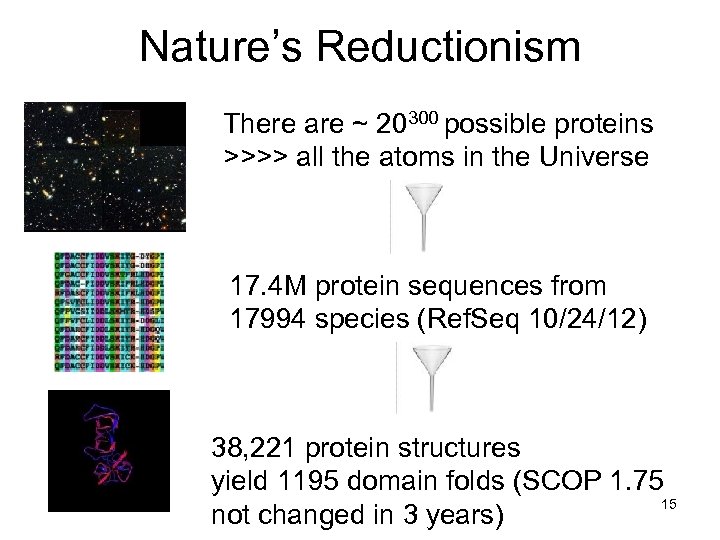 Nature’s Reductionism There are ~ 20300 possible proteins >>>> all the atoms in the Universe 17. 4 M protein sequences from 17994 species (Ref. Seq 10/24/12) 38, 221 protein structures yield 1195 domain folds (SCOP 1. 75 15 not changed in 3 years)
Nature’s Reductionism There are ~ 20300 possible proteins >>>> all the atoms in the Universe 17. 4 M protein sequences from 17994 species (Ref. Seq 10/24/12) 38, 221 protein structures yield 1195 domain folds (SCOP 1. 75 15 not changed in 3 years)
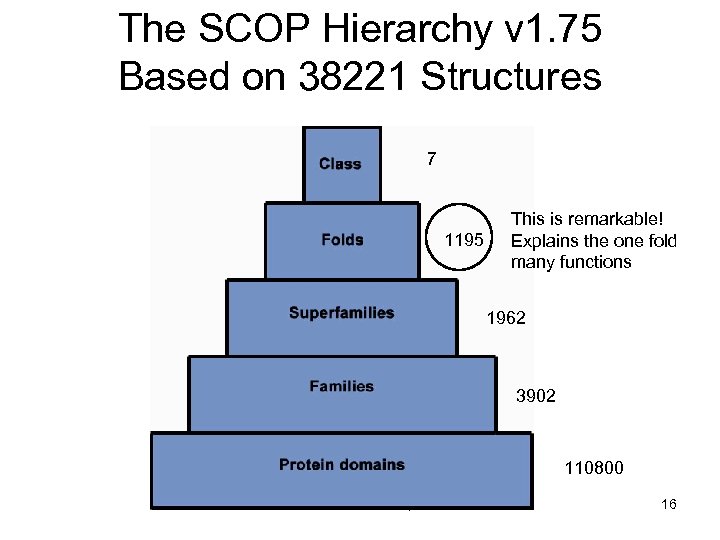 The SCOP Hierarchy v 1. 75 Based on 38221 Structures 7 1195 This is remarkable! Explains the one fold many functions 1962 3902 110800 PHAR 201 Lecture 07, 2012 16
The SCOP Hierarchy v 1. 75 Based on 38221 Structures 7 1195 This is remarkable! Explains the one fold many functions 1962 3902 110800 PHAR 201 Lecture 07, 2012 16
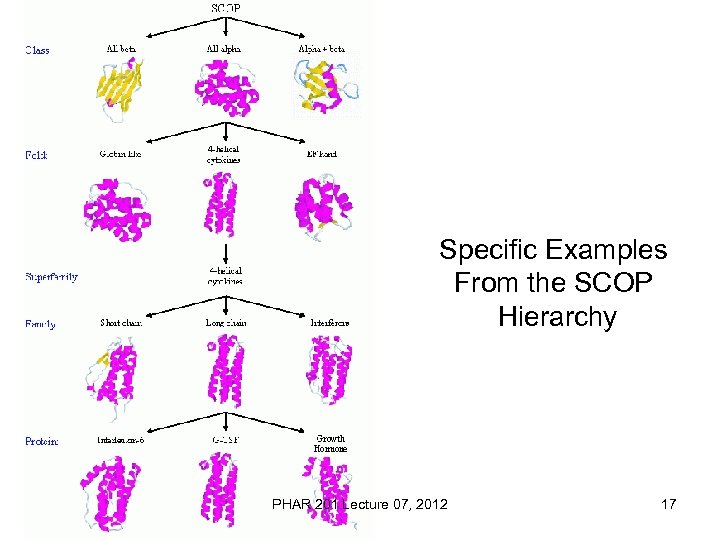 Specific Examples From the SCOP Hierarchy PHAR 201 Lecture 07, 2012 17
Specific Examples From the SCOP Hierarchy PHAR 201 Lecture 07, 2012 17
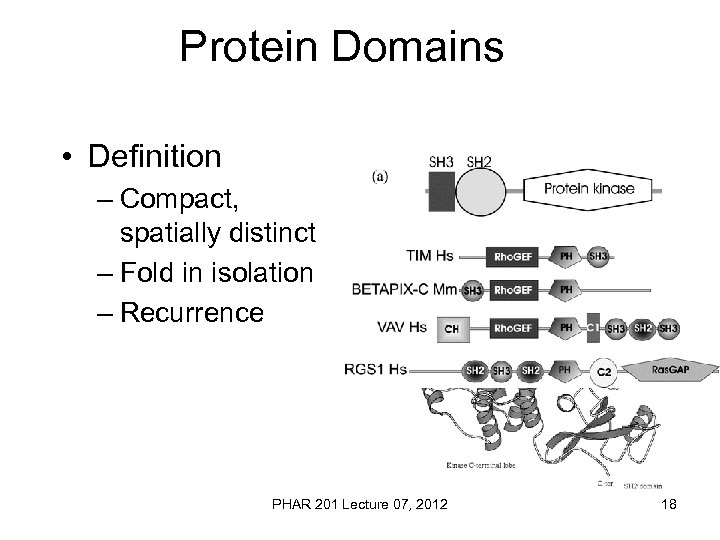 Protein Domains • Definition – Compact, spatially distinct – Fold in isolation – Recurrence PHAR 201 Lecture 07, 2012 18
Protein Domains • Definition – Compact, spatially distinct – Fold in isolation – Recurrence PHAR 201 Lecture 07, 2012 18
 Structure vs Function PHAR 201 Lecture 07, 2012 19
Structure vs Function PHAR 201 Lecture 07, 2012 19
 Some Basic Rules Governing Structure-Function Relationships … • The golden rule is there are no golden rules – George Bernard Shaw • Above 40% sequence identity sequences tend to have the same structure and function – But there are exceptions • Structure and function tend to diverge at the same level of sequence identity PHAR 201 Lecture 07, 2012 20
Some Basic Rules Governing Structure-Function Relationships … • The golden rule is there are no golden rules – George Bernard Shaw • Above 40% sequence identity sequences tend to have the same structure and function – But there are exceptions • Structure and function tend to diverge at the same level of sequence identity PHAR 201 Lecture 07, 2012 20
 Structure vs Function This is even more complicated than the relationship between sequence and structure and not as well understood PHAR 201 Lecture 07, 2012 21
Structure vs Function This is even more complicated than the relationship between sequence and structure and not as well understood PHAR 201 Lecture 07, 2012 21
 Complication Comes from One Structure Multiple Functions • We saw this from GO already • phosphoglucose isomerase acts as a neuroleukin, cytokine and a differentiation mediator as a monomer in the extracellular space and as a dimer in the cell involved in glucose metabolism PHAR 201 Lecture 07, 2012 22
Complication Comes from One Structure Multiple Functions • We saw this from GO already • phosphoglucose isomerase acts as a neuroleukin, cytokine and a differentiation mediator as a monomer in the extracellular space and as a dimer in the cell involved in glucose metabolism PHAR 201 Lecture 07, 2012 22
 Consider an Example Relative to SCOP • lysozyme and alpha-lactalbumin: – Same class alpha+beta – Same superfamily – lysozyme-like – Same family C-type lysozyme – Same fold – lysozyme-like – different function at 40% sequence identity • Lysozyme – hydrolase EC 3. 2. 1. 17 • Alpha lactalbumin – Ca binding lactose biosynthesis PHAR 201 Lecture 07, 2012 23
Consider an Example Relative to SCOP • lysozyme and alpha-lactalbumin: – Same class alpha+beta – Same superfamily – lysozyme-like – Same family C-type lysozyme – Same fold – lysozyme-like – different function at 40% sequence identity • Lysozyme – hydrolase EC 3. 2. 1. 17 • Alpha lactalbumin – Ca binding lactose biosynthesis PHAR 201 Lecture 07, 2012 23
 More Details… Lysozyme is an O-glycosyl hydrolase, but -lactalbumin does not have this catalytic activity. Instead it regulates the substrate specificity of galactosyl transferase through its sugar binding site, which is common to both lactalbumin and lysozyme. Both the sugar binding site and catalytic residues have been retained by lysozyme during evolution, but in -lactalbumin, the catalytic residues have changed and it is no longer an enzyme. PHAR 201 Lecture 07, 2012 24
More Details… Lysozyme is an O-glycosyl hydrolase, but -lactalbumin does not have this catalytic activity. Instead it regulates the substrate specificity of galactosyl transferase through its sugar binding site, which is common to both lactalbumin and lysozyme. Both the sugar binding site and catalytic residues have been retained by lysozyme during evolution, but in -lactalbumin, the catalytic residues have changed and it is no longer an enzyme. PHAR 201 Lecture 07, 2012 24
 Why is It Not so Well Understood? 1. Function is often ill-defined e. g. , biochemical, biological, phonotypical and instances are buried in the literature 2. The PDB is biased – it does not have a balanced repertoire of functions and those functions are ill-defined 3. There a number of functional classifications eg EC, GO that have differing coverage and depth PHAR 201 Lecture 07, 2012 25
Why is It Not so Well Understood? 1. Function is often ill-defined e. g. , biochemical, biological, phonotypical and instances are buried in the literature 2. The PDB is biased – it does not have a balanced repertoire of functions and those functions are ill-defined 3. There a number of functional classifications eg EC, GO that have differing coverage and depth PHAR 201 Lecture 07, 2012 25
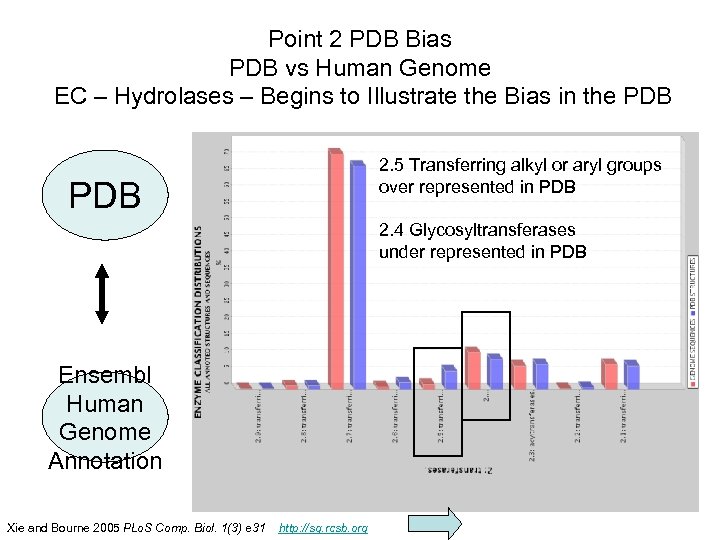 Point 2 PDB Bias PDB vs Human Genome EC – Hydrolases – Begins to Illustrate the Bias in the PDB 2. 5 Transferring alkyl or aryl groups over represented in PDB 2. 4 Glycosyltransferases under represented in PDB Ensembl Human Genome Annotation PHAR 201 Lecture 07, 2012 Xie and Bourne 2005 PLo. S Comp. Biol. 1(3) e 31 http: //sg. rcsb. org 26
Point 2 PDB Bias PDB vs Human Genome EC – Hydrolases – Begins to Illustrate the Bias in the PDB 2. 5 Transferring alkyl or aryl groups over represented in PDB 2. 4 Glycosyltransferases under represented in PDB Ensembl Human Genome Annotation PHAR 201 Lecture 07, 2012 Xie and Bourne 2005 PLo. S Comp. Biol. 1(3) e 31 http: //sg. rcsb. org 26
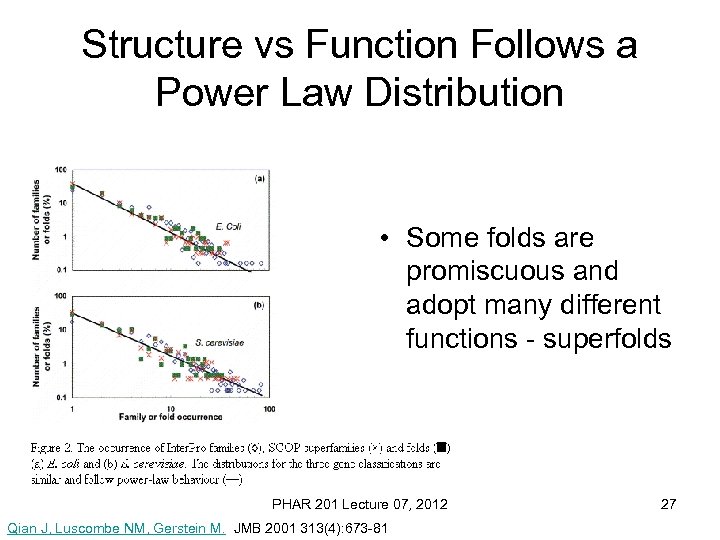 Structure vs Function Follows a Power Law Distribution • Some folds are promiscuous and adopt many different functions - superfolds PHAR 201 Lecture 07, 2012 Qian J, Luscombe NM, Gerstein M. JMB 2001 313(4): 673 -81 27
Structure vs Function Follows a Power Law Distribution • Some folds are promiscuous and adopt many different functions - superfolds PHAR 201 Lecture 07, 2012 Qian J, Luscombe NM, Gerstein M. JMB 2001 313(4): 673 -81 27
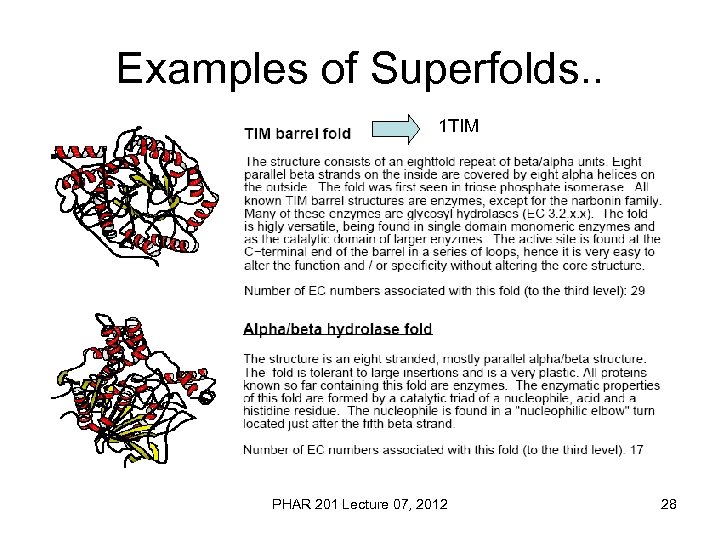 Examples of Superfolds. . 1 TIM PHAR 201 Lecture 07, 2012 28
Examples of Superfolds. . 1 TIM PHAR 201 Lecture 07, 2012 28
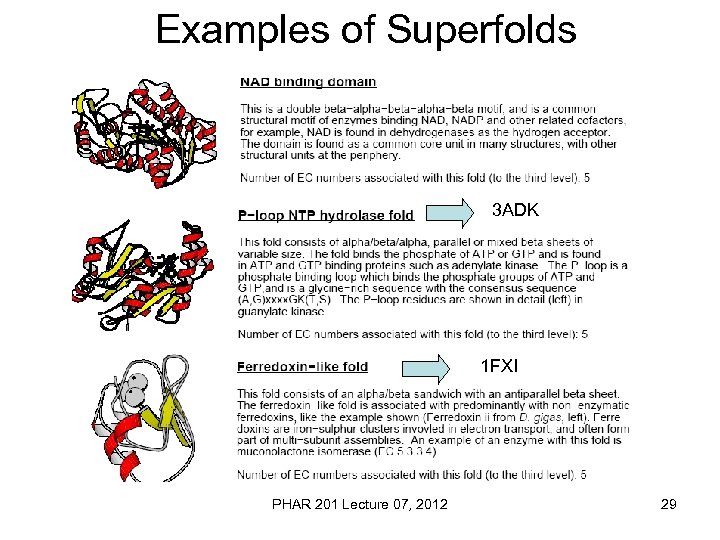 Examples of Superfolds 3 ADK 1 FXI PHAR 201 Lecture 07, 2012 29
Examples of Superfolds 3 ADK 1 FXI PHAR 201 Lecture 07, 2012 29
 Specific Examples of the Relationship Between Structure and Function PHAR 201 Lecture 07, 2012 30
Specific Examples of the Relationship Between Structure and Function PHAR 201 Lecture 07, 2012 30
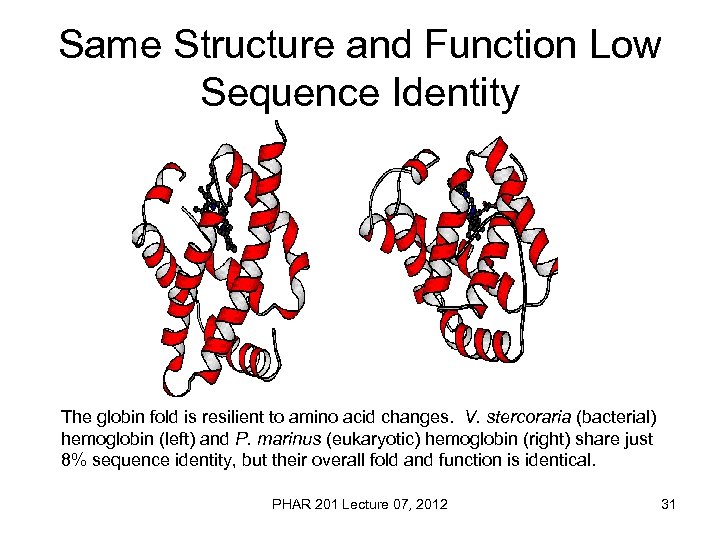 Same Structure and Function Low Sequence Identity The globin fold is resilient to amino acid changes. V. stercoraria (bacterial) hemoglobin (left) and P. marinus (eukaryotic) hemoglobin (right) share just 8% sequence identity, but their overall fold and function is identical. PHAR 201 Lecture 07, 2012 31
Same Structure and Function Low Sequence Identity The globin fold is resilient to amino acid changes. V. stercoraria (bacterial) hemoglobin (left) and P. marinus (eukaryotic) hemoglobin (right) share just 8% sequence identity, but their overall fold and function is identical. PHAR 201 Lecture 07, 2012 31
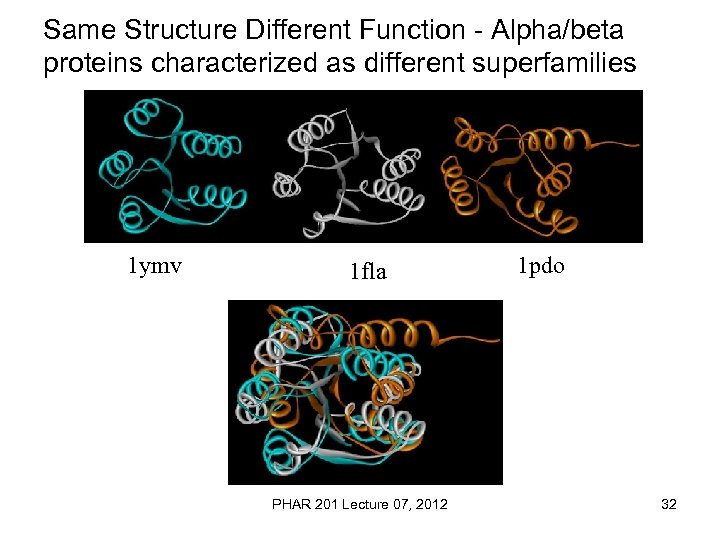 Same Structure Different Function - Alpha/beta proteins characterized as different superfamilies 1 ymv 1 fla PHAR 201 Lecture 07, 2012 1 pdo 32
Same Structure Different Function - Alpha/beta proteins characterized as different superfamilies 1 ymv 1 fla PHAR 201 Lecture 07, 2012 1 pdo 32
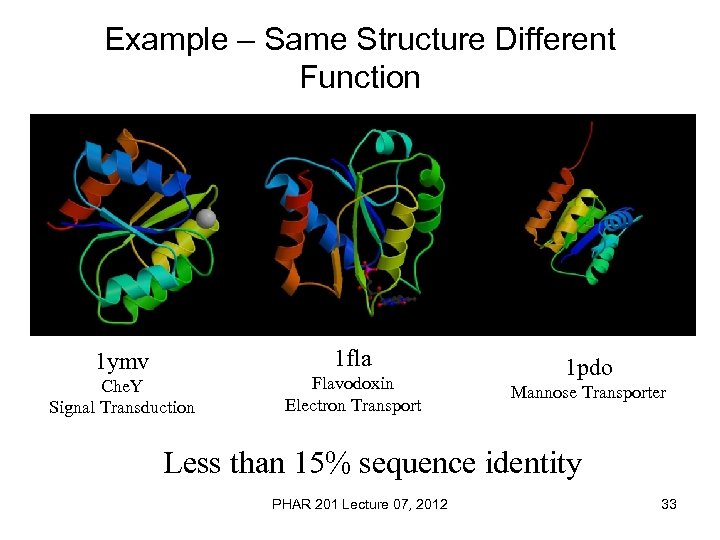 Example – Same Structure Different Function 1 fla 1 ymv Che. Y Signal Transduction Flavodoxin Electron Transport 1 pdo Mannose Transporter Less than 15% sequence identity PHAR 201 Lecture 07, 2012 33
Example – Same Structure Different Function 1 fla 1 ymv Che. Y Signal Transduction Flavodoxin Electron Transport 1 pdo Mannose Transporter Less than 15% sequence identity PHAR 201 Lecture 07, 2012 33
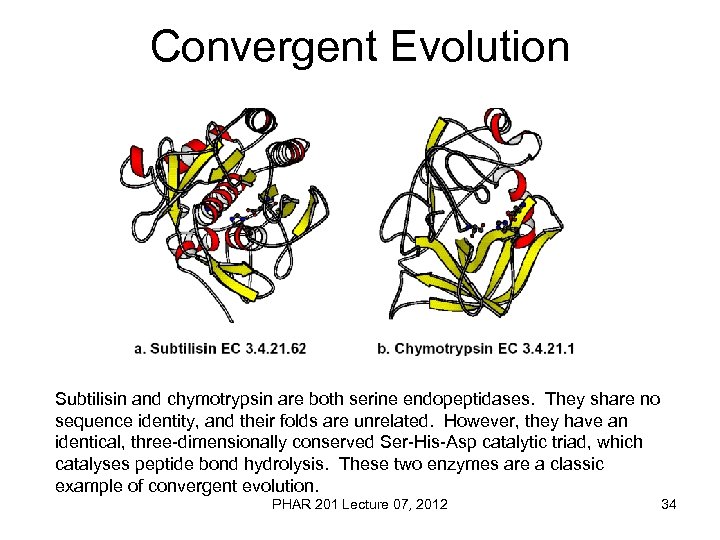 Convergent Evolution Subtilisin and chymotrypsin are both serine endopeptidases. They share no sequence identity, and their folds are unrelated. However, they have an identical, three-dimensionally conserved Ser-His-Asp catalytic triad, which catalyses peptide bond hydrolysis. These two enzymes are a classic example of convergent evolution. PHAR 201 Lecture 07, 2012 34
Convergent Evolution Subtilisin and chymotrypsin are both serine endopeptidases. They share no sequence identity, and their folds are unrelated. However, they have an identical, three-dimensionally conserved Ser-His-Asp catalytic triad, which catalyses peptide bond hydrolysis. These two enzymes are a classic example of convergent evolution. PHAR 201 Lecture 07, 2012 34
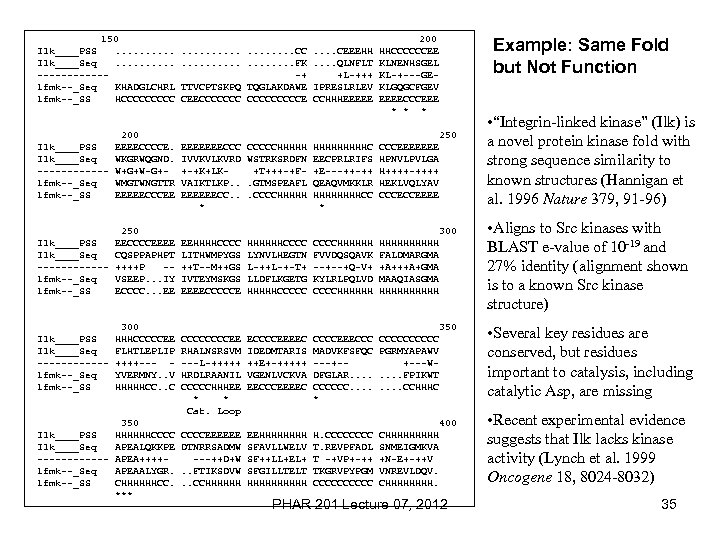 150 Ilk____PSS. . Ilk____Seq. . ------1 fmk--_Seq KHADGLCHRL 1 fmk--_SS HCCCCC 200 Ilk____PSS EEEECCCCE. Ilk____Seq WKGRWQGND. ------ W+G+W-G+1 fmk--_Seq WMGTWNGTTR 1 fmk--_SS EEEEECCCEE Ilk____PSS Ilk____Seq ------1 fmk--_Seq 1 fmk--_SS 250 EECCCCEEEE CQSPPAPHPT ++++P -VSEEP. . . IY ECCCC. . . EE Ilk____PSS Ilk____Seq ------1 fmk--_Seq 1 fmk--_SS 300 HHHCCCCCEE FLHTLEPLIP ++++--- YVERMNY. . V HHHHHCC. . C Ilk____PSS Ilk____Seq ------1 fmk--_Seq 1 fmk--_SS 350 HHHHHHCCCC APEALQKKPE APEA++++APEAALYGR. CHHHHHHCC. *** . . . . CC. . CEEEHH. . . . FK. . QLNFLT -+ +L-+++ TTVCPTSKPQ TQGLAKDAWE IPRESLRLEV CEECCCCCCCE CCHHHEEEEE 200 HHCCCCCCEE KLNENHSGEL KL-+---GEKLGQGCFGEV EEEECCCEEE * * * Example: Same Fold but Not Function 250 EEEEEEECCC IVVKVLKVRD +-+K+LKVAIKTLKP. . EEEEEECC. . * CCCCCHHHHH WSTRKSRDFN +T+++-+F. GTMSPEAFL. CCCCHHHHHC EECPRLRIFS +E---++-++ QEAQVMKKLR HHHHCC * CCCEEEEEEE HPNVLPVLGA H++++-++++ HEKLVQLYAV CCCECCEEEE EEHHHHCCCC LITHWMPYGS ++T--M++GS IVTEYMSKGS EEEECCCCCE HHHHHHCCCC LYNVLHEGTN L-++L-+-T+ LLDFLKGETG HHHHHCCCCCHHHHHH FVVDQSQAVK --+--+Q-V+ KYLRLPQLVD CCCCHHHHHH FALDMARGMA +A+++A+GMA MAAQIASGMA HHHHH CCCCEE RHALNSRSVM ---L-+++++ HRDLRAANIL CCCCCHHHEE * * Cat. Loop ECCCCEEEEC IDEDMTARIS ++E+-+++++ VGENLVCKVA EECCCEEEEC CCCCEEECCC MADVKFSFQC ---+-DFGLAR. . CCCCCC. . * CCCCC PGRMYAPAWV +---W. . FPIKWT. . CCHHHC CCCCEEEEEE DTNRRSADMW ---++D+W. . FTIKSDVW. . CCHHHHHH EEHHHH SFAVLLWELV SF++LL+EL+ SFGILLTELT HHHHH H. CCCC T. REVPFADL T -+VP+-++ TKGRVPYPGM CCCCC CHHHHH SNMEIGMKVA +N-E+-++V VNREVLDQV. CHHHH. 300 350 400 PHAR 201 Lecture 07, 2012 • “Integrin-linked kinase” (Ilk) is a novel protein kinase fold with strong sequence similarity to known structures (Hannigan et al. 1996 Nature 379, 91 -96) • Aligns to Src kinases with BLAST e-value of 10 -19 and 27% identity (alignment shown is to a known Src kinase structure) • Several key residues are conserved, but residues important to catalysis, including catalytic Asp, are missing • Recent experimental evidence suggests that Ilk lacks kinase activity (Lynch et al. 1999 Oncogene 18, 8024 -8032) 35
150 Ilk____PSS. . Ilk____Seq. . ------1 fmk--_Seq KHADGLCHRL 1 fmk--_SS HCCCCC 200 Ilk____PSS EEEECCCCE. Ilk____Seq WKGRWQGND. ------ W+G+W-G+1 fmk--_Seq WMGTWNGTTR 1 fmk--_SS EEEEECCCEE Ilk____PSS Ilk____Seq ------1 fmk--_Seq 1 fmk--_SS 250 EECCCCEEEE CQSPPAPHPT ++++P -VSEEP. . . IY ECCCC. . . EE Ilk____PSS Ilk____Seq ------1 fmk--_Seq 1 fmk--_SS 300 HHHCCCCCEE FLHTLEPLIP ++++--- YVERMNY. . V HHHHHCC. . C Ilk____PSS Ilk____Seq ------1 fmk--_Seq 1 fmk--_SS 350 HHHHHHCCCC APEALQKKPE APEA++++APEAALYGR. CHHHHHHCC. *** . . . . CC. . CEEEHH. . . . FK. . QLNFLT -+ +L-+++ TTVCPTSKPQ TQGLAKDAWE IPRESLRLEV CEECCCCCCCE CCHHHEEEEE 200 HHCCCCCCEE KLNENHSGEL KL-+---GEKLGQGCFGEV EEEECCCEEE * * * Example: Same Fold but Not Function 250 EEEEEEECCC IVVKVLKVRD +-+K+LKVAIKTLKP. . EEEEEECC. . * CCCCCHHHHH WSTRKSRDFN +T+++-+F. GTMSPEAFL. CCCCHHHHHC EECPRLRIFS +E---++-++ QEAQVMKKLR HHHHCC * CCCEEEEEEE HPNVLPVLGA H++++-++++ HEKLVQLYAV CCCECCEEEE EEHHHHCCCC LITHWMPYGS ++T--M++GS IVTEYMSKGS EEEECCCCCE HHHHHHCCCC LYNVLHEGTN L-++L-+-T+ LLDFLKGETG HHHHHCCCCCHHHHHH FVVDQSQAVK --+--+Q-V+ KYLRLPQLVD CCCCHHHHHH FALDMARGMA +A+++A+GMA MAAQIASGMA HHHHH CCCCEE RHALNSRSVM ---L-+++++ HRDLRAANIL CCCCCHHHEE * * Cat. Loop ECCCCEEEEC IDEDMTARIS ++E+-+++++ VGENLVCKVA EECCCEEEEC CCCCEEECCC MADVKFSFQC ---+-DFGLAR. . CCCCCC. . * CCCCC PGRMYAPAWV +---W. . FPIKWT. . CCHHHC CCCCEEEEEE DTNRRSADMW ---++D+W. . FTIKSDVW. . CCHHHHHH EEHHHH SFAVLLWELV SF++LL+EL+ SFGILLTELT HHHHH H. CCCC T. REVPFADL T -+VP+-++ TKGRVPYPGM CCCCC CHHHHH SNMEIGMKVA +N-E+-++V VNREVLDQV. CHHHH. 300 350 400 PHAR 201 Lecture 07, 2012 • “Integrin-linked kinase” (Ilk) is a novel protein kinase fold with strong sequence similarity to known structures (Hannigan et al. 1996 Nature 379, 91 -96) • Aligns to Src kinases with BLAST e-value of 10 -19 and 27% identity (alignment shown is to a known Src kinase structure) • Several key residues are conserved, but residues important to catalysis, including catalytic Asp, are missing • Recent experimental evidence suggests that Ilk lacks kinase activity (Lynch et al. 1999 Oncogene 18, 8024 -8032) 35
 Non-Redundant Sets: Sequences • Refseq (NCBI) – Annotated • BLASTclust http: //www. ncbi. nlm. nih. gov/Web/Newsltr/ Spring 04/blastlab. html • CDhit http: //bioinformatics. org/cd-hit/ popular algorithm for fast clustering of sequences PHAR 201 Lecture 07, 2012 36
Non-Redundant Sets: Sequences • Refseq (NCBI) – Annotated • BLASTclust http: //www. ncbi. nlm. nih. gov/Web/Newsltr/ Spring 04/blastlab. html • CDhit http: //bioinformatics. org/cd-hit/ popular algorithm for fast clustering of sequences PHAR 201 Lecture 07, 2012 36
 Non-Redundant Sets: Sequences with Structure • PDBselect - http: //bioinfo. tg. fhgiessen. de/pdbselect/ • Astral http: //astral. berkeley. edu/ • Pisces http: //dunbrack. fccc. edu/Guoli/PISCES_O ption. Page. php • RCSB PDB queries • RCSB Sequence Similaity PHAR 201 Lecture 07, 2012 37
Non-Redundant Sets: Sequences with Structure • PDBselect - http: //bioinfo. tg. fhgiessen. de/pdbselect/ • Astral http: //astral. berkeley. edu/ • Pisces http: //dunbrack. fccc. edu/Guoli/PISCES_O ption. Page. php • RCSB PDB queries • RCSB Sequence Similaity PHAR 201 Lecture 07, 2012 37
 PHAR 201 Lecture 07, 2012 38
PHAR 201 Lecture 07, 2012 38
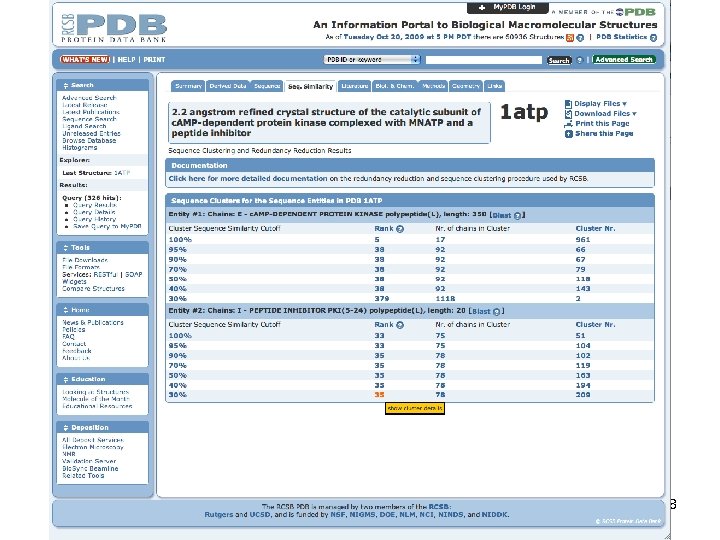
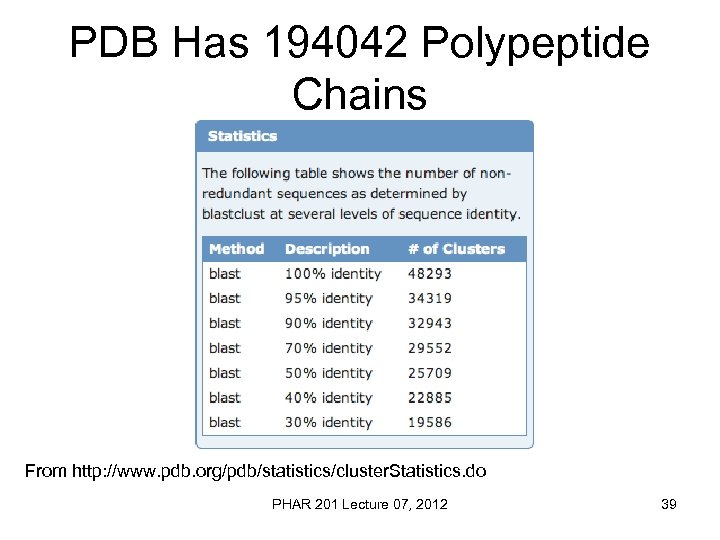 PDB Has 194042 Polypeptide Chains From http: //www. pdb. org/pdb/statistics/cluster. Statistics. do PHAR 201 Lecture 07, 2012 39
PDB Has 194042 Polypeptide Chains From http: //www. pdb. org/pdb/statistics/cluster. Statistics. do PHAR 201 Lecture 07, 2012 39


