3ad0678d84092b02f612fe0e46726cd6.ppt
- Количество слайдов: 38

Tutorial: Bioinformatics Resources (http: //pir. georgetown. edu/~huz/class/bioinfo_resource. html) Bio-Trac 25 (Proteomics: Principles and Methods) March 26, 2004 Zhang-Zhi Hu, M. D. Senior Bioinformatics Scientist Protein Information Resource National Biomedical Research Foundation, GUMC
What is Bioinformatics? computer + mouse = bioinformatics (information) (biology) NIH Biomedical Information Science and Technology Initiative (BISTI) Working Definition (2000) - Research, development, or application of computational tools and approaches for expanding the use of biological, medical, behavioral or health data, including those to acquire, store, organize, archive, analyze, or visualize such data. 2
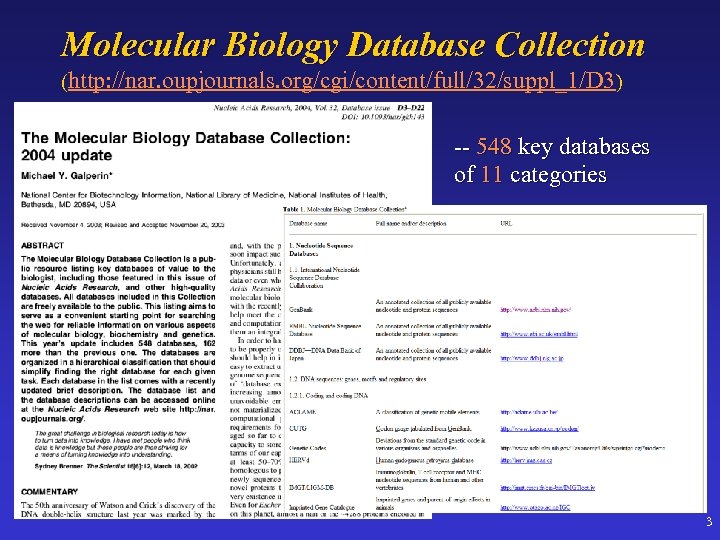
Molecular Biology Database Collection (http: //nar. oupjournals. org/cgi/content/full/32/suppl_1/D 3) -- 548 key databases of 11 categories 3

(http: //pir. georgetown. edu/~huz/class/2004_database_update. html) 4

Overview Database Contents, Search and Retrieval I. III. IV. V. VII. Text search / Information retrieval Sequence & genomics databases Protein family databases Database of protein functions Databases of protein structures 2 D-gel databases Proteomics databases 5
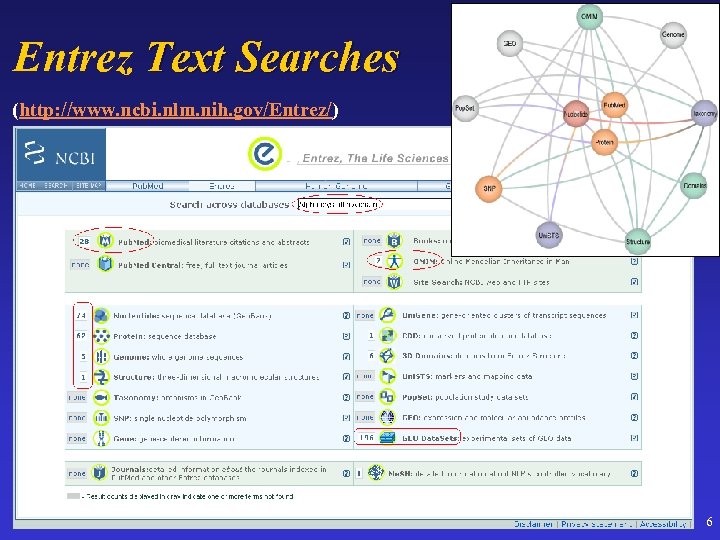
Entrez Text Searches (http: //www. ncbi. nlm. nih. gov/Entrez/) 6
Pub. Med Literature Database (http: //www. ncbi. nlm. nih. gov/entrez/query. fcgi? CMD=Search&DB=Pub. Med) 7
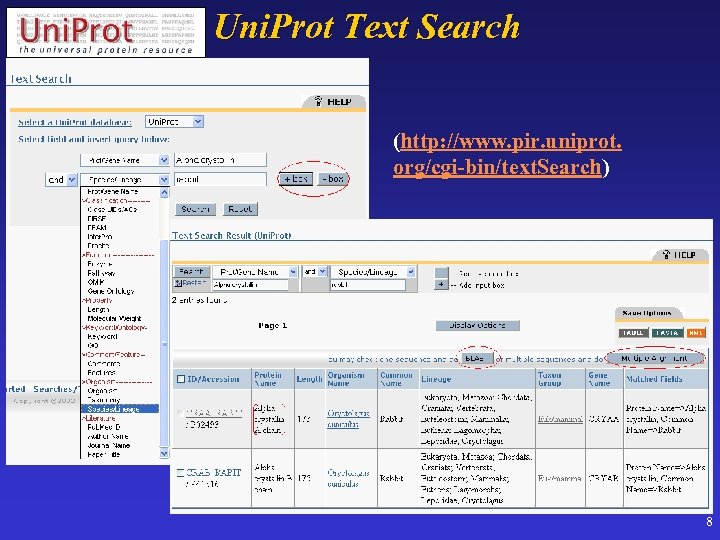
Uni. Prot Text Search (http: //www. pir. uniprot. org/cgi-bin/text. Search) 8
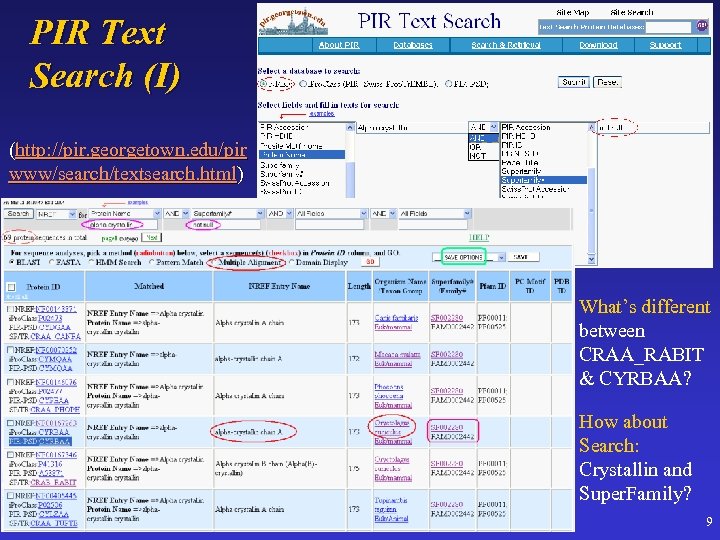
PIR Text Search (I) (http: //pir. georgetown. edu/pir www/search/textsearch. html) What’s different between CRAA_RABIT & CYRBAA? How about Search: Crystallin and Super. Family? 9
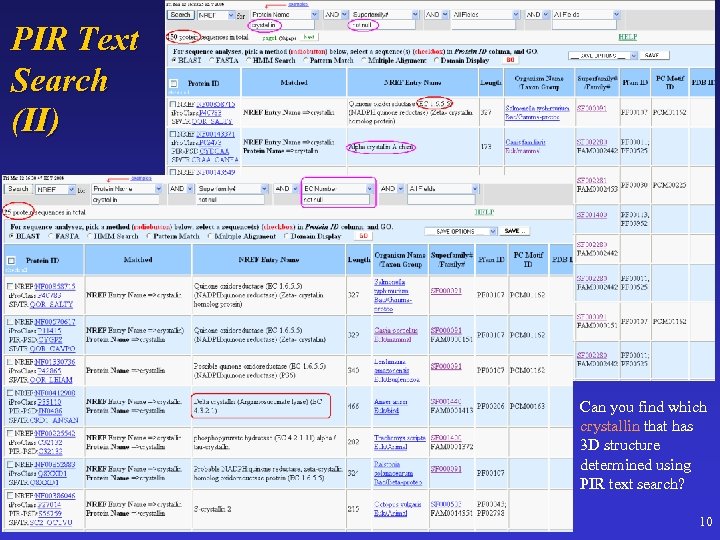
PIR Text Search (II) Can you find which crystallin that has 3 D structure determined using PIR text search? 10
I. Sequence & Genomics Databases Gen. Bank: An annotated collection of all publicly available nucleotide and protein sequences. Ref. Seq: NCBI non-redundant set of reference sequences, including genomic DNA, transcript (RNA), and protein products Uni. Prot Consortium Database: Universal protein knowledgebase, a central resource of protein sequence and function from Swiss-Prot, Tr. EMBL and PIR. Locus. Link: Curated sequences and descriptions of genetic loci. Uni. Gene: Unified clusters of ESTs and full-length m. RNA sequences. OMIM: Online Mendelian inheritance in man: a catalog of human genetic and genomic disorders. Model Organism Genome Databases: MGD, RGD, SGD, Flybase… Gene. Cards: Integrated database of human genes, maps, proteins and diseases. SNP Consortium Database 11
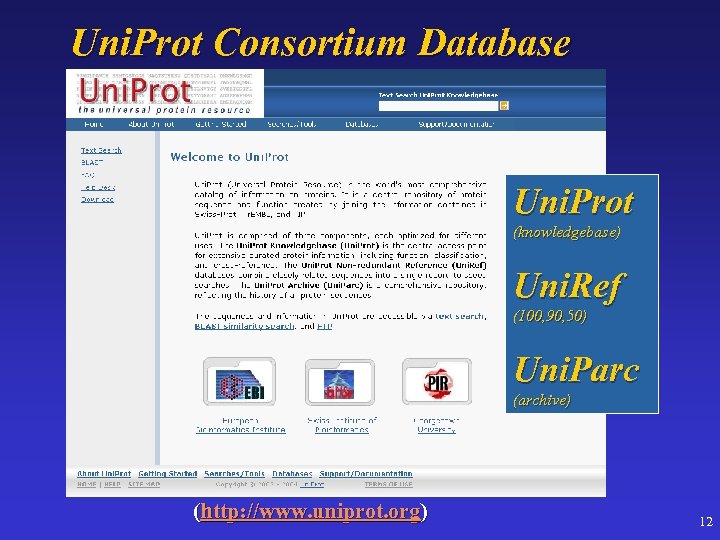
Uni. Prot Consortium Database Uni. Prot (knowledgebase) Uni. Ref (100, 90, 50) Uni. Parc (archive) (http: //www. uniprot. org) 12
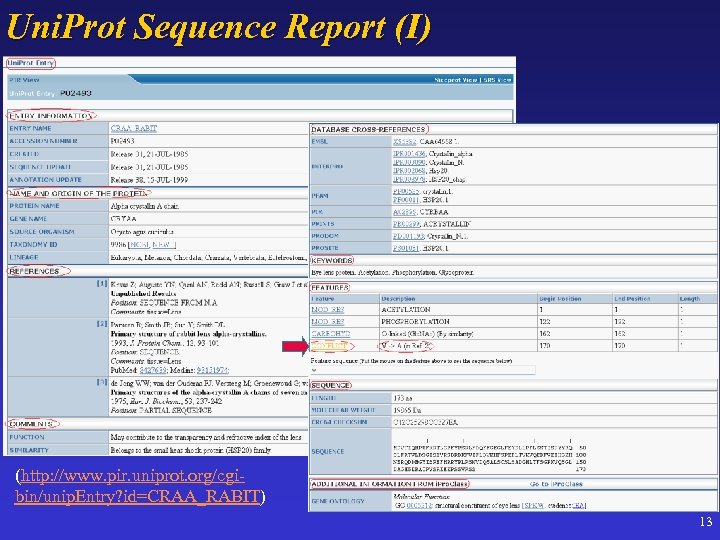
Uni. Prot Sequence Report (I) (http: //www. pir. uniprot. org/cgibin/unip. Entry? id=CRAA_RABIT) 13
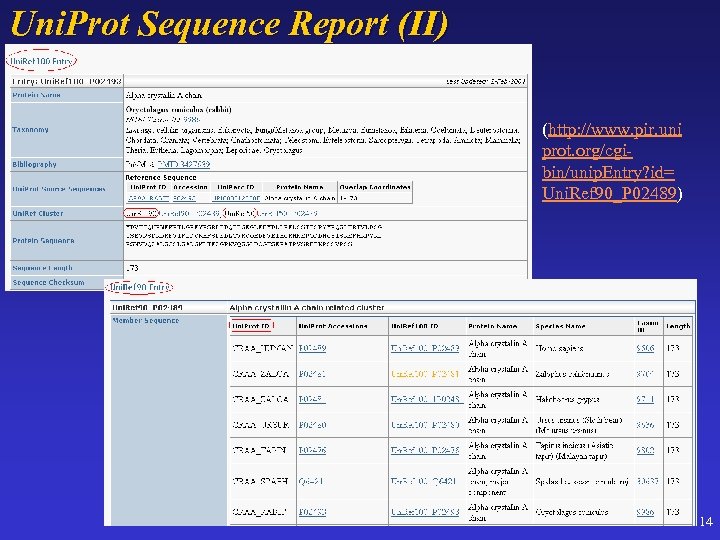
Uni. Prot Sequence Report (II) (http: //www. pir. uni prot. org/cgibin/unip. Entry? id= Uni. Ref 90_P 02489) 14

NCBI Locus. Link (http: //www. ncbi. nlm. nih. gov/Locus. Link) 15
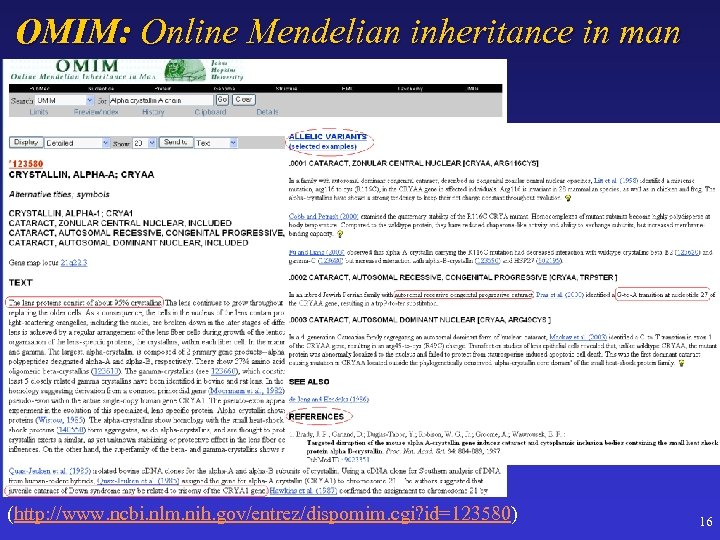
OMIM: Online Mendelian inheritance in man (http: //www. ncbi. nlm. nih. gov/entrez/dispomim. cgi? id=123580) 16
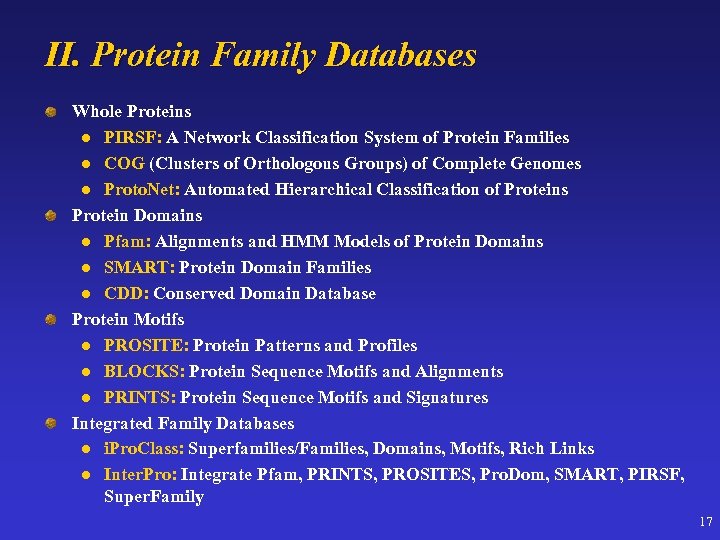
II. Protein Family Databases Whole Proteins l PIRSF: A Network Classification System of Protein Families l COG (Clusters of Orthologous Groups) of Complete Genomes l Proto. Net: Automated Hierarchical Classification of Proteins Protein Domains l Pfam: Alignments and HMM Models of Protein Domains l SMART: Protein Domain Families l CDD: Conserved Domain Database Protein Motifs l PROSITE: Protein Patterns and Profiles l BLOCKS: Protein Sequence Motifs and Alignments l PRINTS: Protein Sequence Motifs and Signatures Integrated Family Databases l i. Pro. Class: Superfamilies/Families, Domains, Motifs, Rich Links l Inter. Pro: Integrate Pfam, PRINTS, PROSITES, Pro. Dom, SMART, PIRSF, Super. Family 17
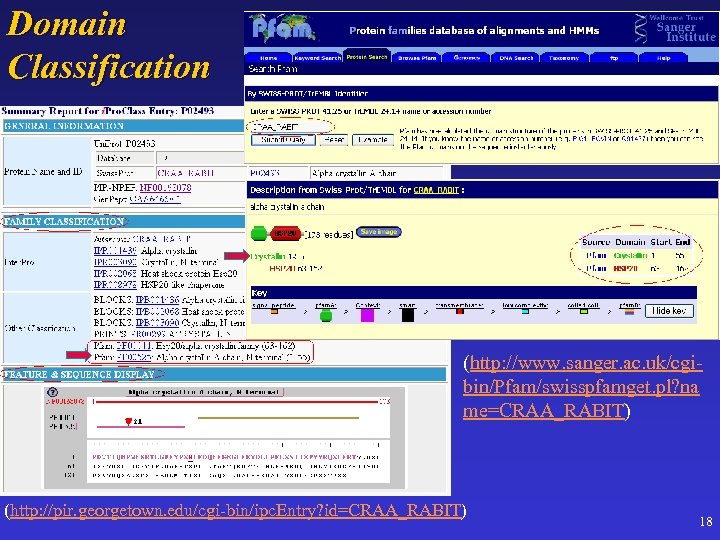
Domain Classification (http: //www. sanger. ac. uk/cgibin/Pfam/swisspfamget. pl? na me=CRAA_RABIT) (http: //pir. georgetown. edu/cgi-bin/ipc. Entry? id=CRAA_RABIT) 18
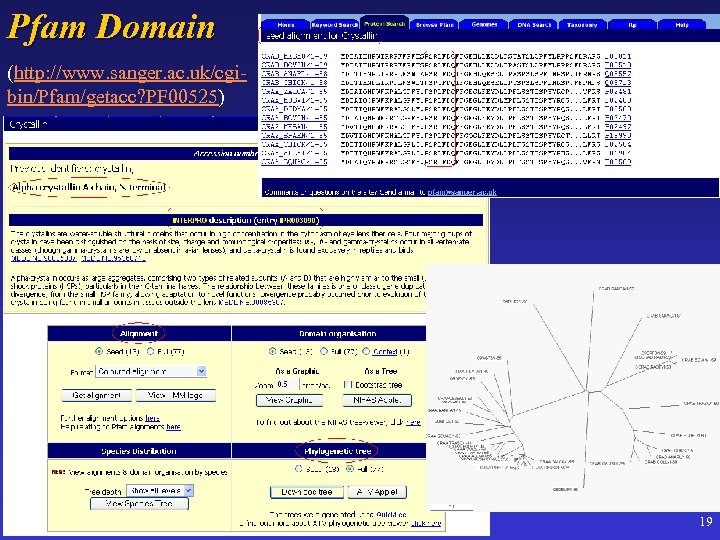
Pfam Domain (http: //www. sanger. ac. uk/cgibin/Pfam/getacc? PF 00525) 19
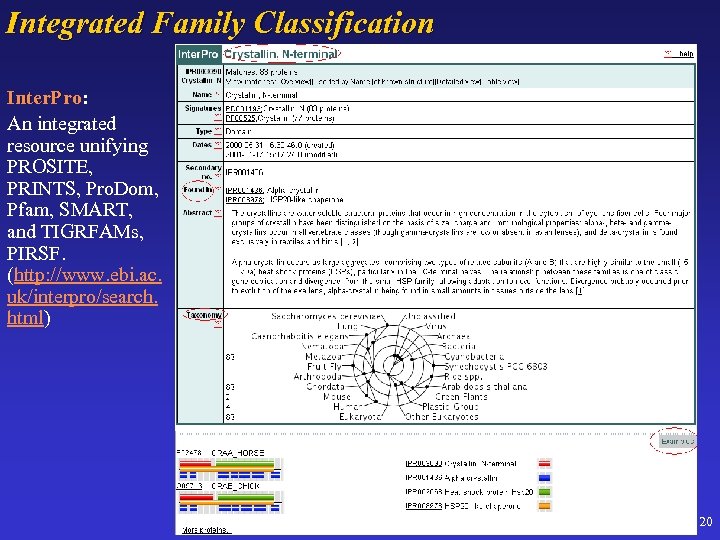
Integrated Family Classification Inter. Pro: Inter. Pro An integrated resource unifying PROSITE, PRINTS, Pro. Dom, Pfam, SMART, and TIGRFAMs, PIRSF. (http: //www. ebi. ac. uk/interpro/search. html) 20
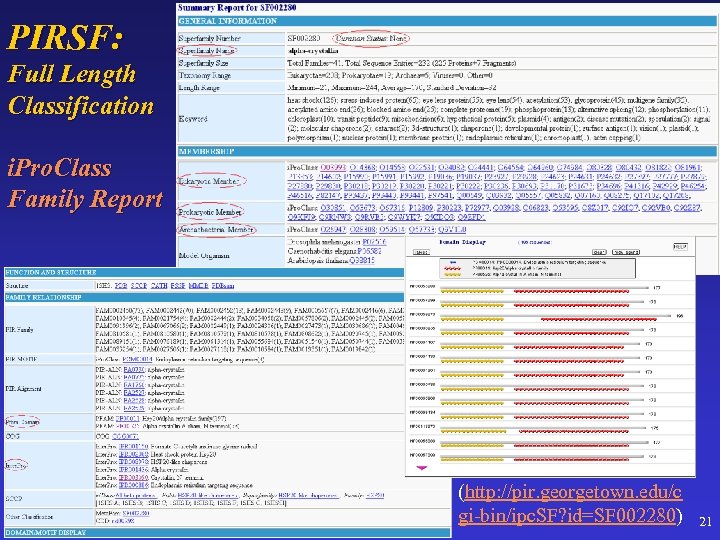
PIRSF: Full Length Classification i. Pro. Class Family Report (http: //pir. georgetown. edu/c gi-bin/ipc. SF? id=SF 002280) 21
III. Databases of Protein Functions Metabolic Pathways, Enzymes, and Compounds l l l l Enzyme Classification: Classification and Nomenclature of Enzyme-Catalysed Reactions (EC-IUBMB) KEGG (Kyoto Encyclopedia of Genes and Genomes): Metabolic Pathways LIGAND (at KEGG): Chemical Compounds, Reactions and Enzymes Eco. Cyc: Encyclopedia of E. coli Genes and Metabolism Meta. Cyc: Metabolic Encyclopedia (Metabolic Pathways) WIT: Functional Curation and Metabolic Models BRENDA: Enzyme Database UM-BBD: Microbial Biocatalytic Reactions and Biodegradation Pathways Cellular Regulation and Gene Networks l l l Epo. DB: Genes Expressed during Human Erythropoiesis BIND: Descriptions of interactions, molecular complexes and pathways DIP: Catalogs experimentally determined interactions between proteins Bio. Carta: Biological pathways of human and mouse GO: Gene Ontology Consortium Database 22
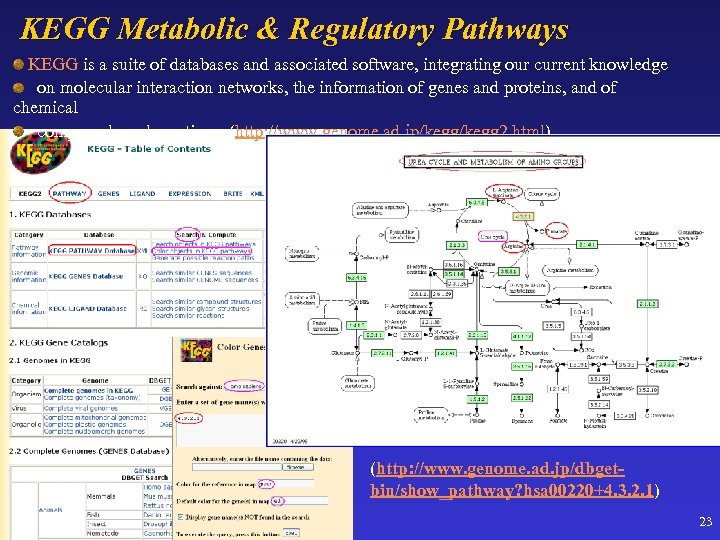
KEGG Metabolic & Regulatory Pathways KEGG is a suite of databases and associated software, integrating our current knowledge on molecular interaction networks, the information of genes and proteins, and of chemical compounds and reactions. (http: //www. genome. ad. jp/kegg 2. html) (http: //www. genome. ad. jp/dbgetbin/show_pathway? hsa 00220+4. 3. 2. 1) 23
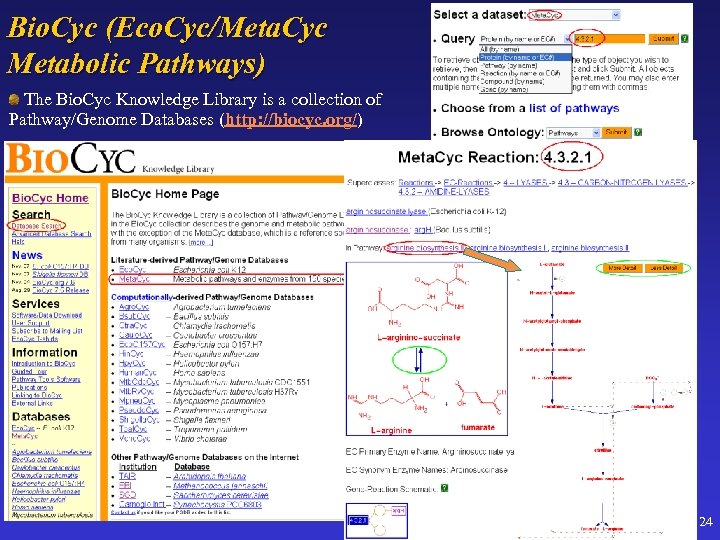
Bio. Cyc (Eco. Cyc/Meta. Cyc Metabolic Pathways) The Bio. Cyc Knowledge Library is a collection of Pathway/Genome Databases (http: //biocyc. org/) 24
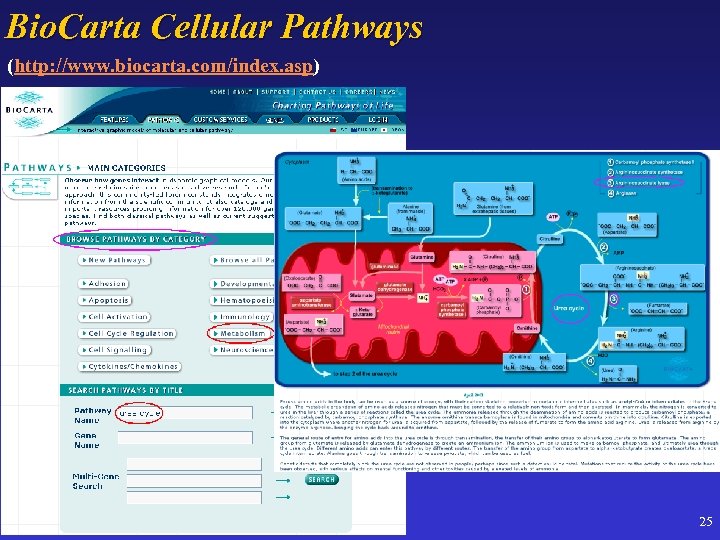
Bio. Carta Cellular Pathways (http: //www. biocarta. com/index. asp) 25
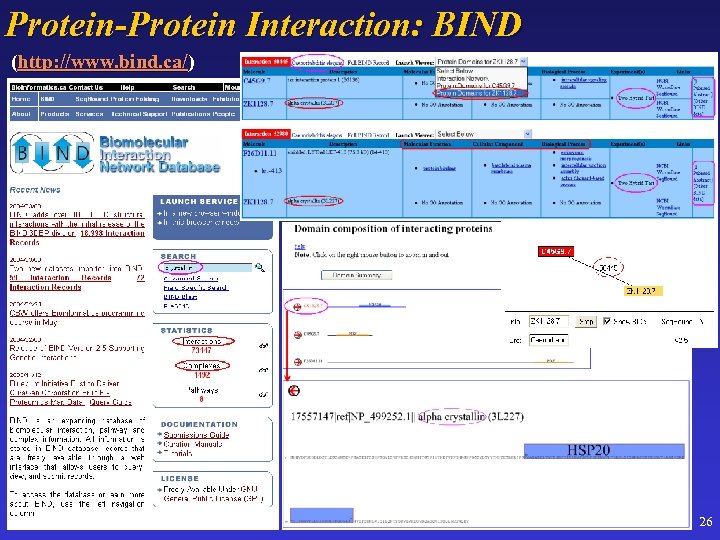
Protein-Protein Interaction: BIND (http: //www. bind. ca/) 26
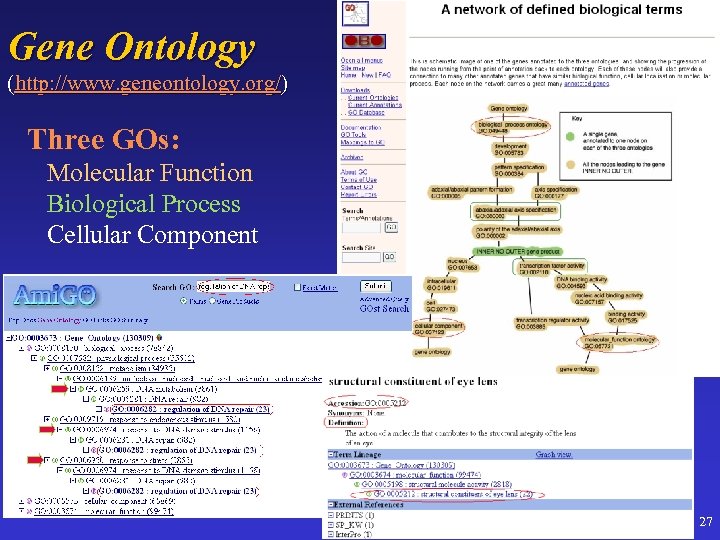
Gene Ontology (http: //www. geneontology. org/) Three GOs: Molecular Function Biological Process Cellular Component 27
IV. Databases of Protein Structures Protein Structure l l l PDB: Structure Determined by X-ray Crystallography and NMR PDBsum: Summaries and analyses of PDB structures MMDB: NCBI’s database of 3 D structures, part of NCBI Entrez SWISS-MODEL Repository: Database of annotated protein 3 D models Mod. Base: Annotated comparative protein structure models Structure Classification l l l CATH: Hierarchical Classification of Protein Domain Structures SCOP: Familial and Structural Protein Relationships FSSP: Protein Fold Classification Based on Structure--Structure Alignment 28
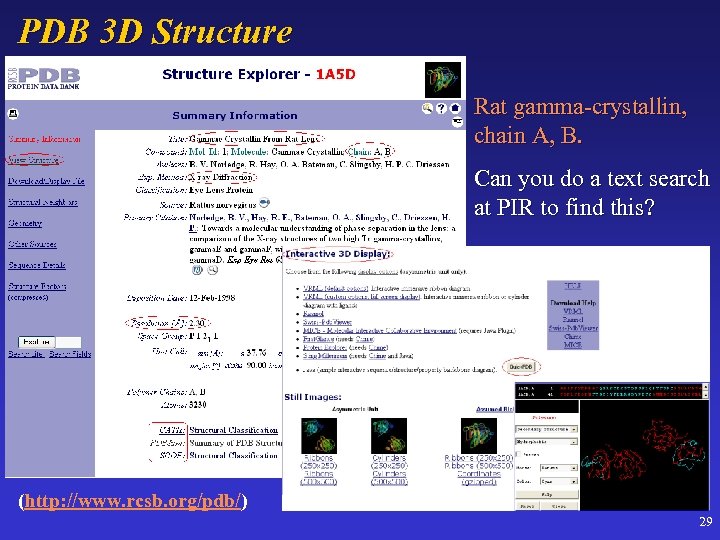
PDB 3 D Structure Rat gamma-crystallin, chain A, B. Can you do a text search at PIR to find this? (http: //www. rcsb. org/pdb/) 29
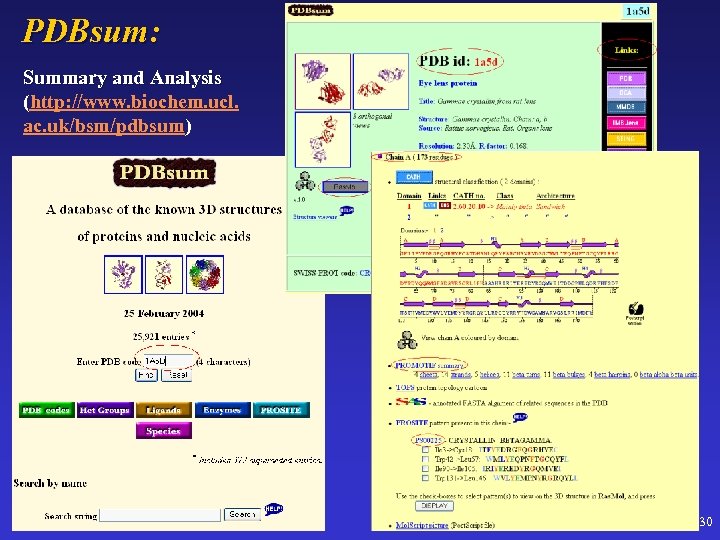
PDBsum: Summary and Analysis (http: //www. biochem. ucl. ac. uk/bsm/pdbsum) 30
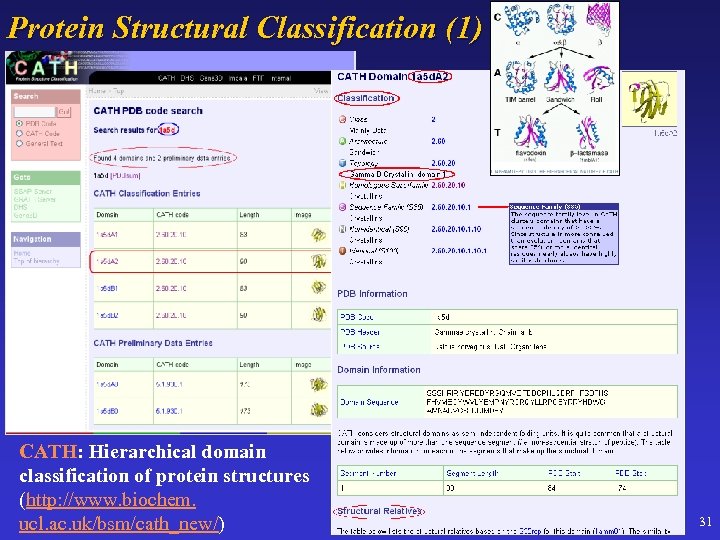
Protein Structural Classification (1) CATH: Hierarchical domain classification of protein structures (http: //www. biochem. ucl. ac. uk/bsm/cath_new/) 31
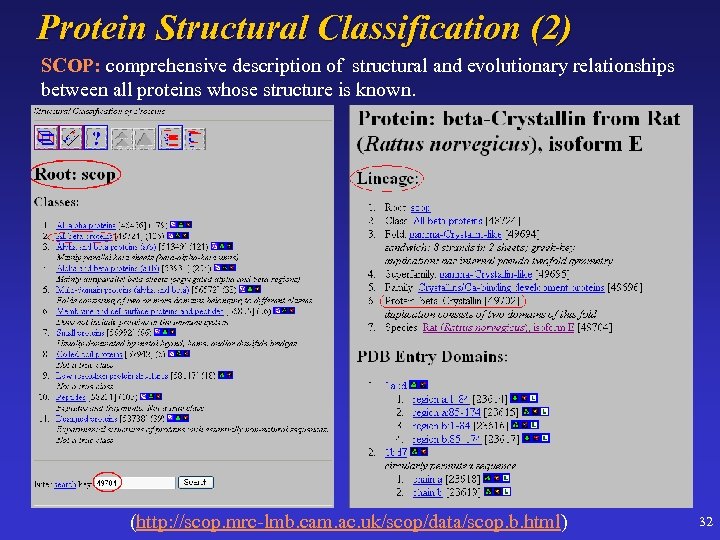
Protein Structural Classification (2) SCOP: comprehensive description of structural and evolutionary relationships between all proteins whose structure is known. (http: //scop. mrc-lmb. cam. ac. uk/scop/data/scop. b. html) 32
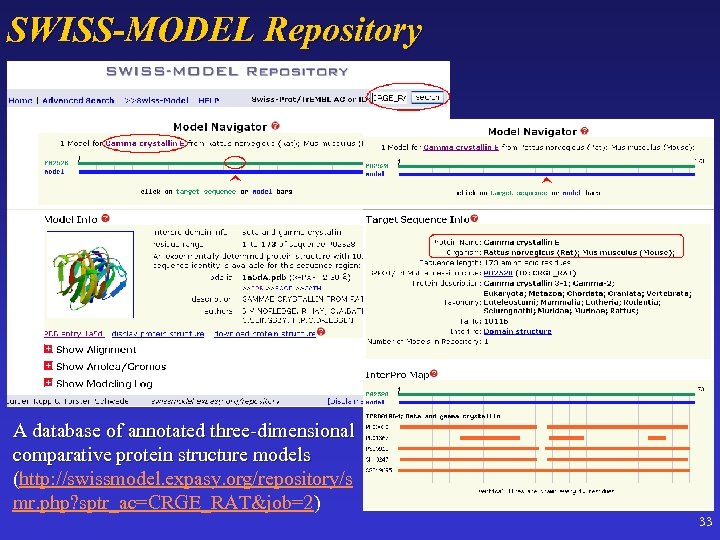
SWISS-MODEL Repository A database of annotated three-dimensional comparative protein structure models (http: //swissmodel. expasy. org/repository/s mr. php? sptr_ac=CRGE_RAT&job=2) 33

VI. Proteomic Resources GELBANK (http: //gelbank. anl. gov): 2 D-gel patterns from completed genomes; SWISS-2 DPAGE (http: //www. expasy. org/ch 2 d/) PEP: Predictions for Entire Proteomes: (http: //cubic. bioc. columbia. edu/ pep/): Summarized analyses of protein sequences Proteome Bio. Knowledge Library: (http: //www. proteome. com): Detailed information on human, mouse and rat proteomes Proteome Analysis Database (http: //www. ebi. ac. uk/proteome/): Online application of Inter. Pro and Clu. STr for the functional classification of proteins in whole genomes Expression Profiling databases: GNF (http: //expression. gnf. org/cgibin/index. cgi, human and mouse transcriptome), SMD (http: //genomewww 5. stanford. edu/Micro. Array/SMD/, Stanford microarray data analysis), EBI Microarray Informatics (http: //www. ebi. ac. uk/microarray/ index. html , managing, storing and analyzing microarray data) 34
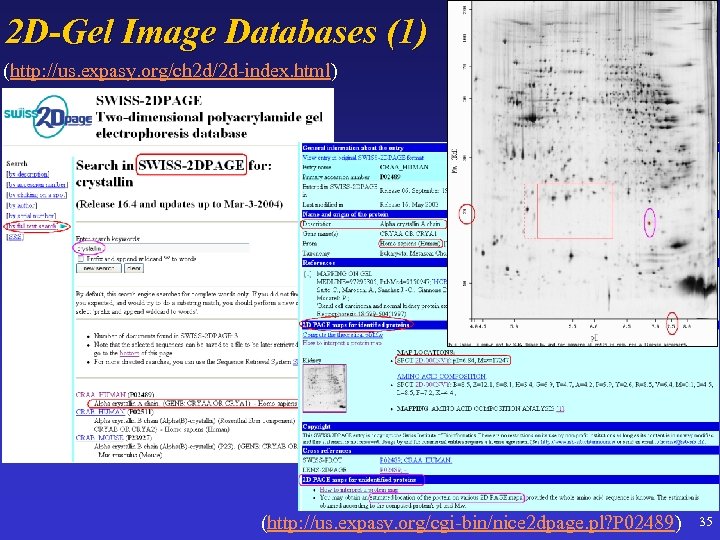
2 D-Gel Image Databases (1) (http: //us. expasy. org/ch 2 d/2 d-index. html) (http: //us. expasy. org/cgi-bin/nice 2 dpage. pl? P 02489) 35
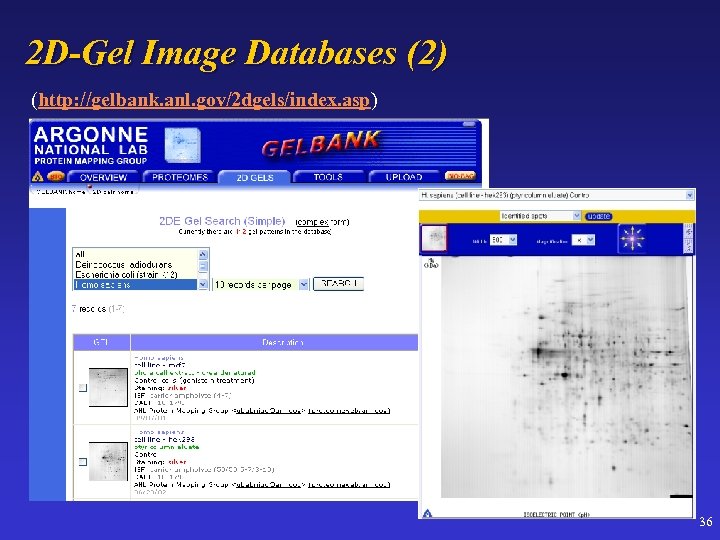
2 D-Gel Image Databases (2) (http: //gelbank. anl. gov/2 dgels/index. asp) 36
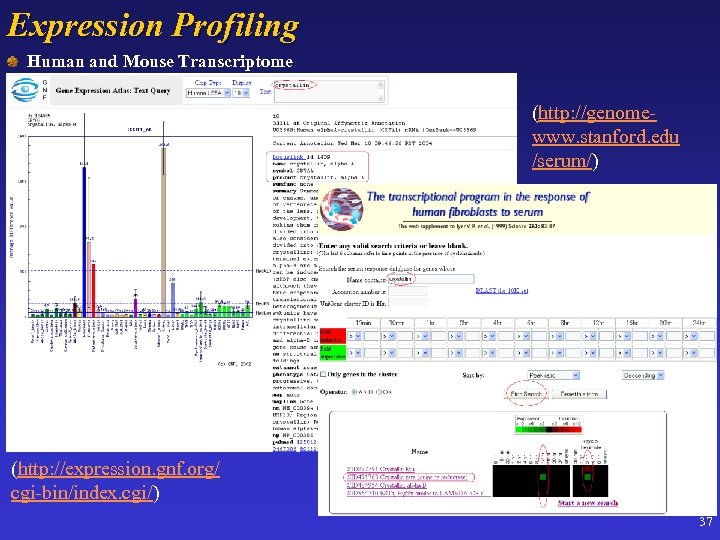
Expression Profiling Human and Mouse Transcriptome (http: //genomewww. stanford. edu /serum/) (http: //expression. gnf. org/cgi-bin/index. cgi) (http: //expression. gnf. org/ cgi-bin/index. cgi/) 37
Lab: Alpha crystallin (Uni. Prot: CRAA_RABIT) Delta crystallin II (Argininosuccinate lyase) (Uni. Prot: CRD 2_ANAPL) Choose additional protein IDs to browse the variety of molecular biology databases each sequence report links to. 38
3ad0678d84092b02f612fe0e46726cd6.ppt