3cee99c85cc84e081bb2f5e075628aa5.ppt
- Количество слайдов: 42
 Towards an intelligent framework to quickly find data from distributed heterogeneous biomedical resources. Despoina Antonakaki, Dasha Zhernakova, Erik Roos, K Joeri van der Velde, Mark Kiestra, Tomasz Adamusiak, Niran Abeygunawardena, Helen Parkinson, Rolf Sijmons, Morris A. Swertz
Towards an intelligent framework to quickly find data from distributed heterogeneous biomedical resources. Despoina Antonakaki, Dasha Zhernakova, Erik Roos, K Joeri van der Velde, Mark Kiestra, Tomasz Adamusiak, Niran Abeygunawardena, Helen Parkinson, Rolf Sijmons, Morris A. Swertz
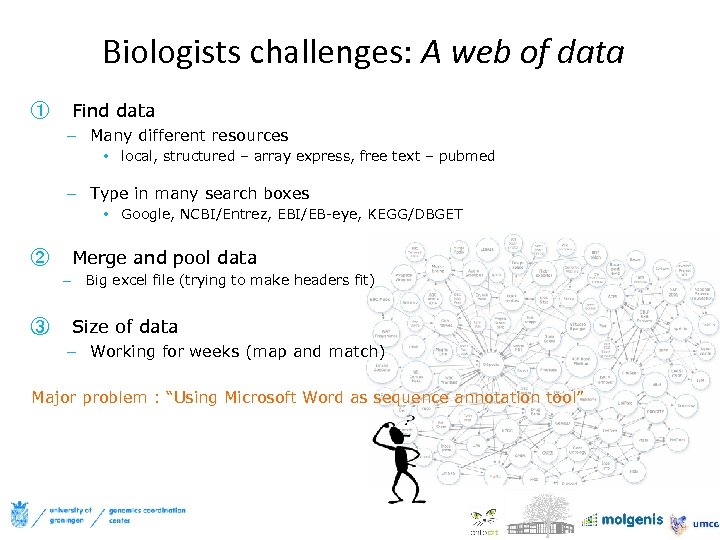 Biologists challenges: A web of data ① Find data – Many different resources • local, structured – array express, free text – pubmed – Type in many search boxes • Google, NCBI/Entrez, EBI/EB-eye, KEGG/DBGET ② Merge and pool data – Big excel file (trying to make headers fit) ③ Size of data – Working for weeks (map and match) Major problem : “Using Microsoft Word as sequence annotation tool”
Biologists challenges: A web of data ① Find data – Many different resources • local, structured – array express, free text – pubmed – Type in many search boxes • Google, NCBI/Entrez, EBI/EB-eye, KEGG/DBGET ② Merge and pool data – Big excel file (trying to make headers fit) ③ Size of data – Working for weeks (map and match) Major problem : “Using Microsoft Word as sequence annotation tool”
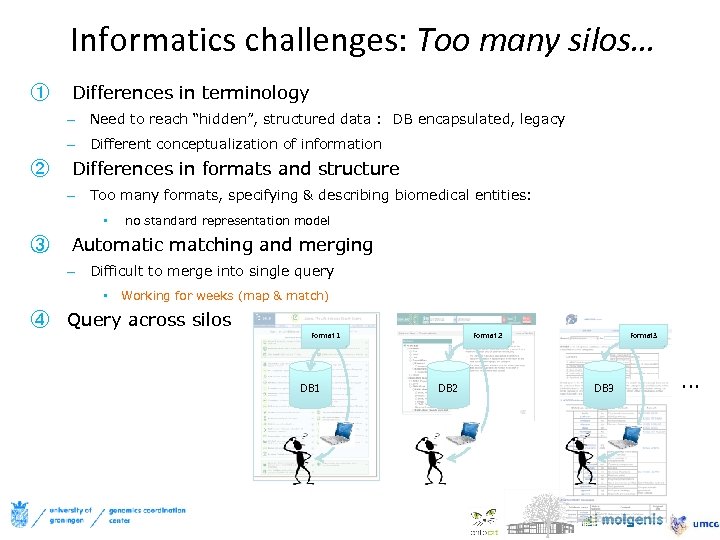 Informatics challenges: Too many silos… ① Differences in terminology – Need to reach “hidden”, structured data : DB encapsulated, legacy – Different conceptualization of information ② Differences in formats and structure – Too many formats, specifying & describing biomedical entities: • ③ no standard representation model Automatic matching and merging – Difficult to merge into single query • Working for weeks (map & match) ④ Query across silos Format 1 DB 1 Format 2 DB 2 Format 3 DB 3 …
Informatics challenges: Too many silos… ① Differences in terminology – Need to reach “hidden”, structured data : DB encapsulated, legacy – Different conceptualization of information ② Differences in formats and structure – Too many formats, specifying & describing biomedical entities: • ③ no standard representation model Automatic matching and merging – Difficult to merge into single query • Working for weeks (map & match) ④ Query across silos Format 1 DB 1 Format 2 DB 2 Format 3 DB 3 …
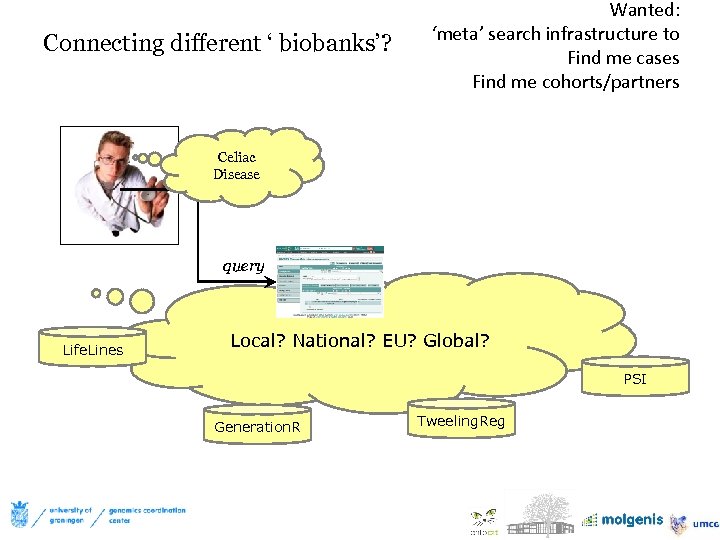 Connecting different ‘ biobanks’? Wanted: ‘meta’ search infrastructure to Find me cases Find me cohorts/partners Celiac Disease query Life. Lines Local? National? EU? Global? PSI Generation. R Tweeling. Reg
Connecting different ‘ biobanks’? Wanted: ‘meta’ search infrastructure to Find me cases Find me cohorts/partners Celiac Disease query Life. Lines Local? National? EU? Global? PSI Generation. R Tweeling. Reg
 Outline • Three challenges for biologists’ and the corresponding for the Informatics’: 1. 2. 3. 4. Merge and pool data - Differences in formats and structure Find data - Differences in terminology Size of data - Automatic matching and merging Across data sets – All above + distribution • Approaches 1. 2. 3. 4. Integrate data into one ‘pheno’ model (MOLGENIS) Use ontologies (Onto. CAT) Indexing (Lucene) Query expansion (Lucene + Onto. CAT) • Discussion 1. Federated data queries (molgenis & rdf)
Outline • Three challenges for biologists’ and the corresponding for the Informatics’: 1. 2. 3. 4. Merge and pool data - Differences in formats and structure Find data - Differences in terminology Size of data - Automatic matching and merging Across data sets – All above + distribution • Approaches 1. 2. 3. 4. Integrate data into one ‘pheno’ model (MOLGENIS) Use ontologies (Onto. CAT) Indexing (Lucene) Query expansion (Lucene + Onto. CAT) • Discussion 1. Federated data queries (molgenis & rdf)
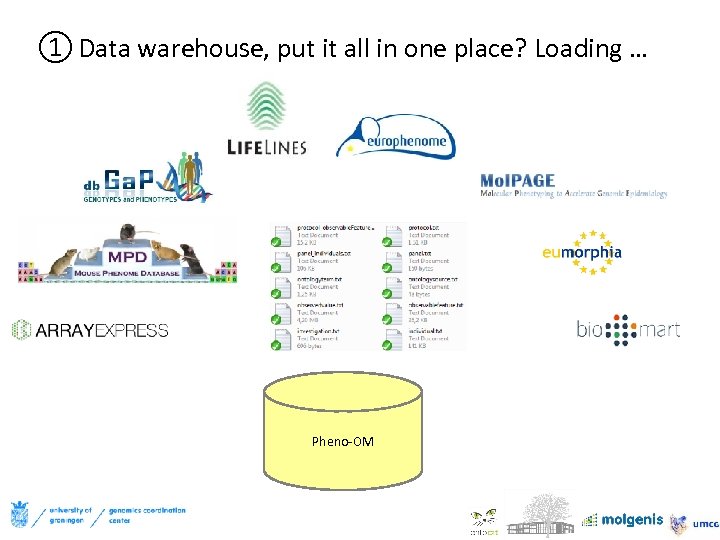 ① Data warehouse, put it all in one place? Loading … Pheno-OM
① Data warehouse, put it all in one place? Loading … Pheno-OM
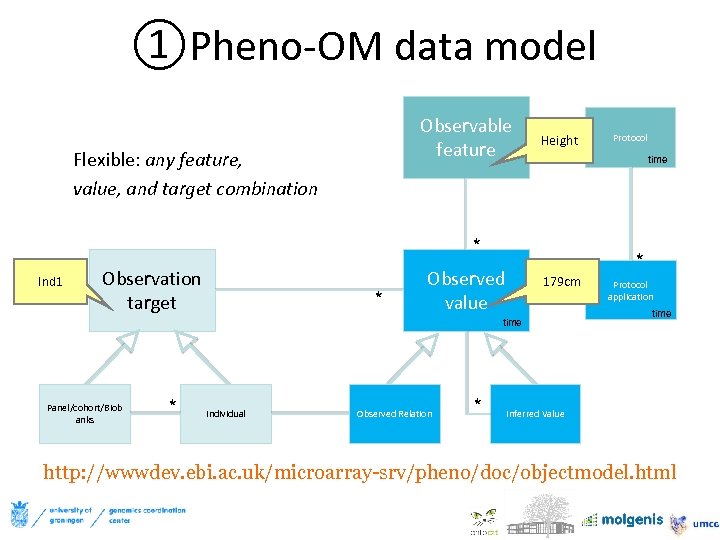 ①Pheno-OM data model Observable * * Height feature Flexible: any feature, value, and target combination * Ind 1 Observation target * * Individual Observed value Observed Relation * time * *179 cm time Panel/cohort/Biob anks Protocol application time Inferred Value http: //wwwdev. ebi. ac. uk/microarray-srv/pheno/doc/objectmodel. html
①Pheno-OM data model Observable * * Height feature Flexible: any feature, value, and target combination * Ind 1 Observation target * * Individual Observed value Observed Relation * time * *179 cm time Panel/cohort/Biob anks Protocol application time Inferred Value http: //wwwdev. ebi. ac. uk/microarray-srv/pheno/doc/objectmodel. html
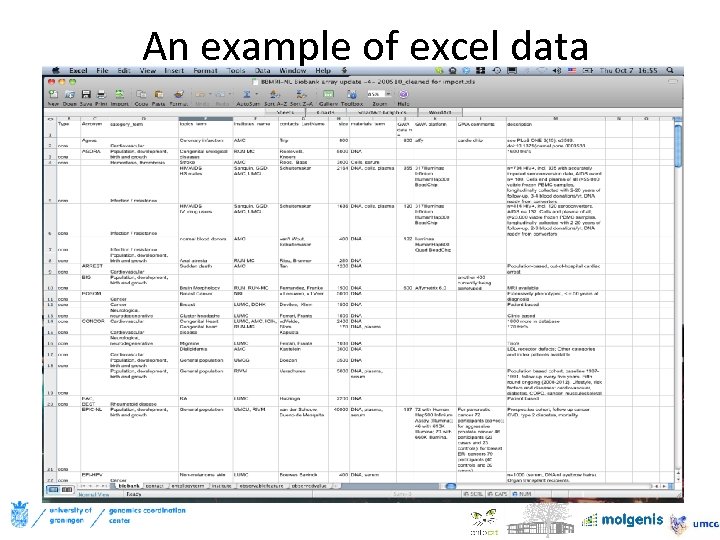 An example of excel data • Or bbmri-nl
An example of excel data • Or bbmri-nl
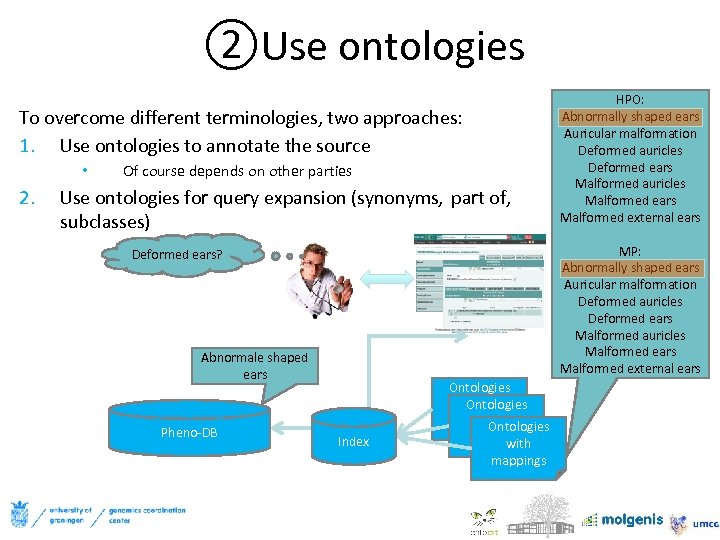 ②Use ontologies To overcome different terminologies, two approaches: 1. Use ontologies to annotate the source • 2. Of course depends on other parties Use ontologies for query expansion (synonyms, part of, subclasses) MP: Abnormally shaped ears Auricular malformation Deformed auricles Deformed ears Malformed auricles Malformed ears Malformed external ears Deformed ears? Abnormale shaped ears Pheno-DB HPO: Abnormally shaped ears Auricular malformation Deformed auricles Deformed ears Malformed auricles Malformed ears Malformed external ears Index Ontologies with Ontologies mappings with mappings
②Use ontologies To overcome different terminologies, two approaches: 1. Use ontologies to annotate the source • 2. Of course depends on other parties Use ontologies for query expansion (synonyms, part of, subclasses) MP: Abnormally shaped ears Auricular malformation Deformed auricles Deformed ears Malformed auricles Malformed ears Malformed external ears Deformed ears? Abnormale shaped ears Pheno-DB HPO: Abnormally shaped ears Auricular malformation Deformed auricles Deformed ears Malformed auricles Malformed ears Malformed external ears Index Ontologies with Ontologies mappings with mappings
 Outline • Three challenges for biologists’ and the corresponding for the Informatics’: 1. 2. 3. 4. Merge and pool data - Differences in formats and structure Find data - Differences in terminology Size of data - Automatic matching and merging Across data sets – All above + distribution • Approaches 1. 2. 3. 4. Integrate data into one ‘pheno’ model (MOLGENIS) Use ontologies (Onto. CAT) Indexing (Lucene) Query expansion (Lucene + Onto. CAT) • Discussion 1. Federated data queries (molgenis & rdf)
Outline • Three challenges for biologists’ and the corresponding for the Informatics’: 1. 2. 3. 4. Merge and pool data - Differences in formats and structure Find data - Differences in terminology Size of data - Automatic matching and merging Across data sets – All above + distribution • Approaches 1. 2. 3. 4. Integrate data into one ‘pheno’ model (MOLGENIS) Use ontologies (Onto. CAT) Indexing (Lucene) Query expansion (Lucene + Onto. CAT) • Discussion 1. Federated data queries (molgenis & rdf)
 Complexity in Ontologies To search across different ontologies requires expert knowledge . . sometimes they change unpredictably. . or sometimes they become suddenly unavailable. .
Complexity in Ontologies To search across different ontologies requires expert knowledge . . sometimes they change unpredictably. . or sometimes they become suddenly unavailable. .
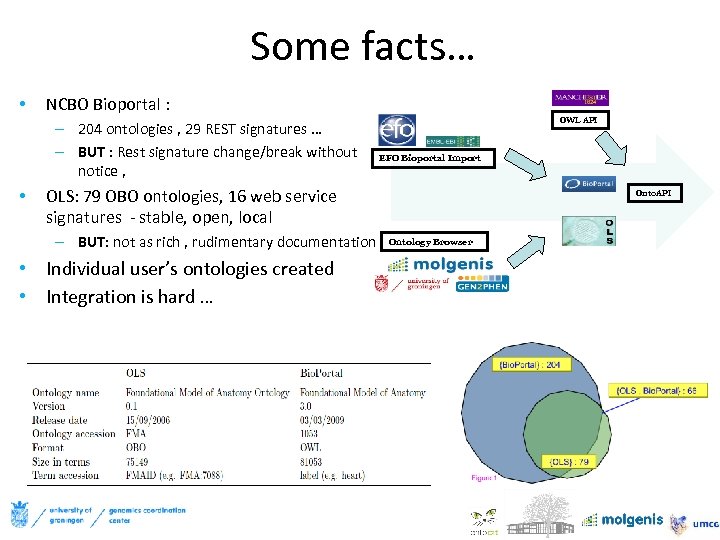 Some facts… • NCBO Bioportal : – 204 ontologies , 29 REST signatures … – BUT : Rest signature change/break without notice , • OWL API EFO Bioportal Import OLS: 79 OBO ontologies, 16 web service signatures - stable, open, local – BUT: not as rich , rudimentary documentation • Individual user’s ontologies created • Integration is hard … Onto. API Ontology Browser
Some facts… • NCBO Bioportal : – 204 ontologies , 29 REST signatures … – BUT : Rest signature change/break without notice , • OWL API EFO Bioportal Import OLS: 79 OBO ontologies, 16 web service signatures - stable, open, local – BUT: not as rich , rudimentary documentation • Individual user’s ontologies created • Integration is hard … Onto. API Ontology Browser
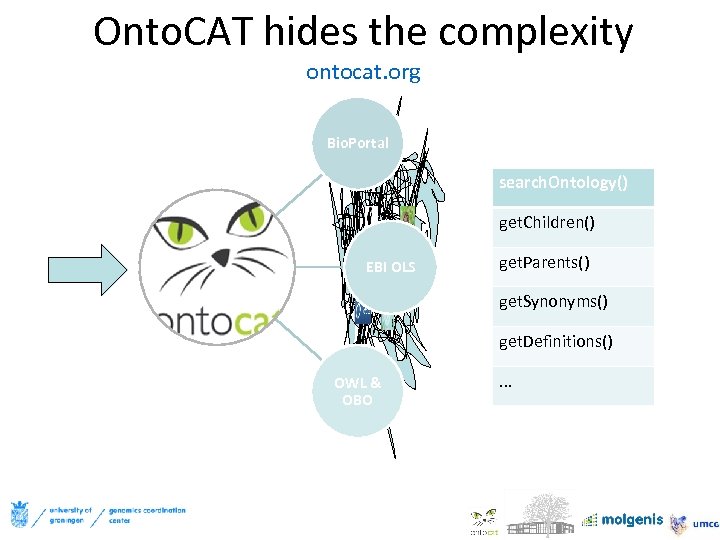 Onto. CAT hides the complexity ontocat. org Bio. Portal search. Ontology() get. Children() EBI OLS get. Parents() get. Synonyms() get. Definitions() OWL & OBO . . .
Onto. CAT hides the complexity ontocat. org Bio. Portal search. Ontology() get. Children() EBI OLS get. Parents() get. Synonyms() get. Definitions() OWL & OBO . . .
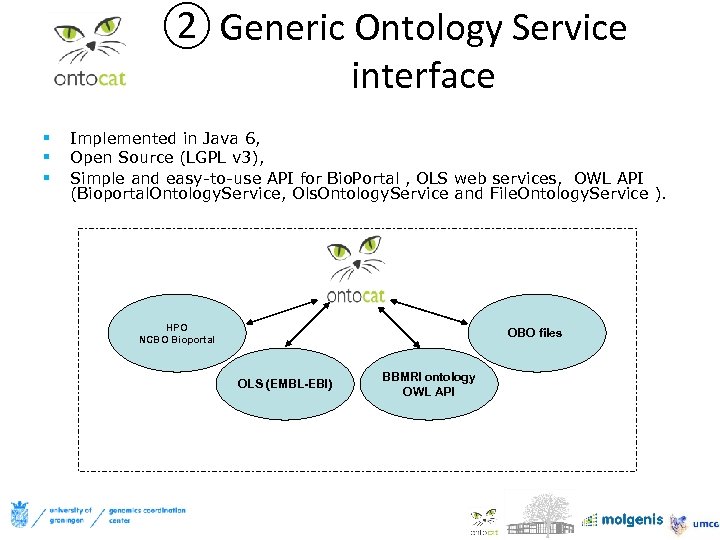 ② Generic Ontology Service interface § § § Implemented in Java 6, Open Source (LGPL v 3), Simple and easy-to-use API for Bio. Portal , OLS web services, OWL API (Bioportal. Ontology. Service, Ols. Ontology. Service and File. Ontology. Service ). HPO NCBO Bioportal OBO files OLS (EMBL-EBI) BBMRI ontology OWL API
② Generic Ontology Service interface § § § Implemented in Java 6, Open Source (LGPL v 3), Simple and easy-to-use API for Bio. Portal , OLS web services, OWL API (Bioportal. Ontology. Service, Ols. Ontology. Service and File. Ontology. Service ). HPO NCBO Bioportal OBO files OLS (EMBL-EBI) BBMRI ontology OWL API
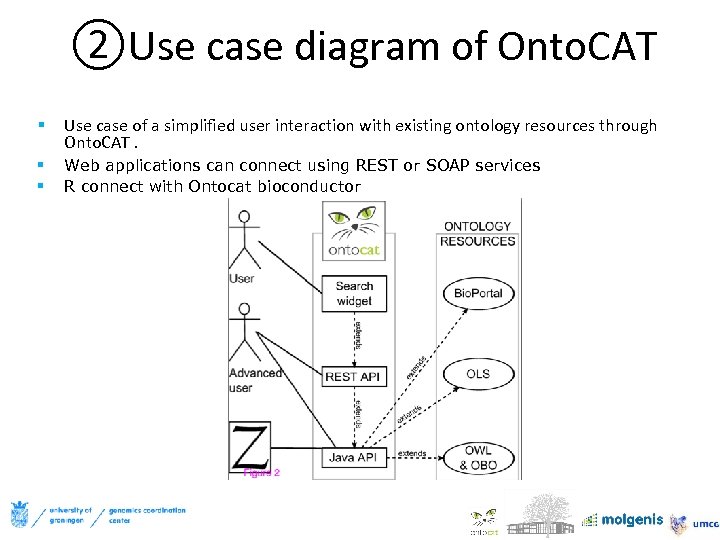 ②Use case diagram of Onto. CAT § Use case of a simplified user interaction with existing ontology resources through Onto. CAT. § § Web applications can connect using REST or SOAP services R connect with Ontocat bioconductor
②Use case diagram of Onto. CAT § Use case of a simplified user interaction with existing ontology resources through Onto. CAT. § § Web applications can connect using REST or SOAP services R connect with Ontocat bioconductor
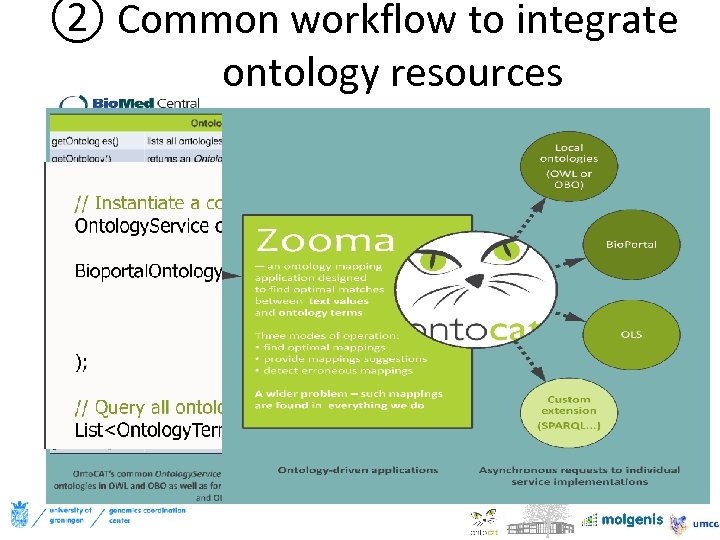 ② Common workflow to integrate ontology resources
② Common workflow to integrate ontology resources
 ② Ontocat example : Find “membrane” term in multiple ontologies
② Ontocat example : Find “membrane” term in multiple ontologies
 ②More examples available
②More examples available
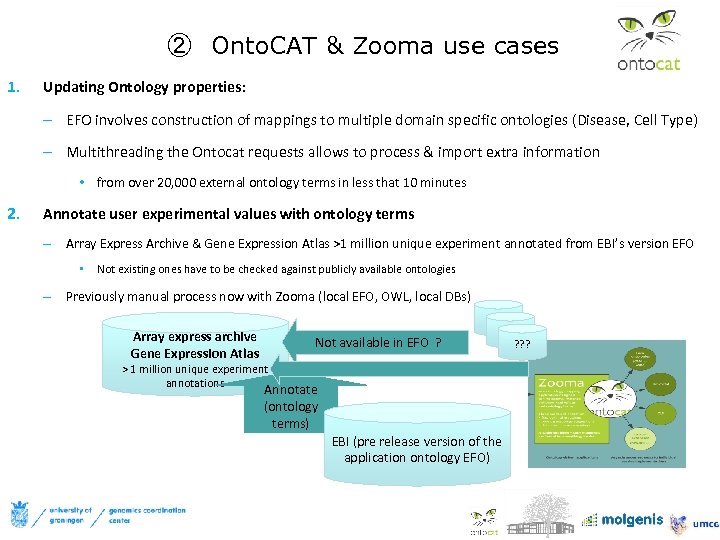 ② Onto. CAT & Zooma use cases 1. Updating Ontology properties: – EFO involves construction of mappings to multiple domain specific ontologies (Disease, Cell Type) – Multithreading the Ontocat requests allows to process & import extra information • from over 20, 000 external ontology terms in less that 10 minutes 2. Annotate user experimental values with ontology terms – Array Express Archive & Gene Expression Atlas >1 million unique experiment annotated from EBI’s version EFO • Not existing ones have to be checked against publicly available ontologies – Previously manual process now with Zooma (local EFO, OWL, local DBs) Array express archive Gene Expression Atlas Not available in EFO ? ? ? ? ? ? > 1 million unique experiment annotations Annotate (ontology terms) EBI (pre release version of the application ontology EFO)
② Onto. CAT & Zooma use cases 1. Updating Ontology properties: – EFO involves construction of mappings to multiple domain specific ontologies (Disease, Cell Type) – Multithreading the Ontocat requests allows to process & import extra information • from over 20, 000 external ontology terms in less that 10 minutes 2. Annotate user experimental values with ontology terms – Array Express Archive & Gene Expression Atlas >1 million unique experiment annotated from EBI’s version EFO • Not existing ones have to be checked against publicly available ontologies – Previously manual process now with Zooma (local EFO, OWL, local DBs) Array express archive Gene Expression Atlas Not available in EFO ? ? ? ? ? ? > 1 million unique experiment annotations Annotate (ontology terms) EBI (pre release version of the application ontology EFO)
 ② Onto. CAT & Zooma use cases 3. Local ontology management – e. Xtensive Genotype And Phenotype data platform (XGAP - Molgenis) : search widget Interactive annotation of data with ontology terms • 4. Allows search publically available ontologies & download terms for unambiguous annotation of QTL or GWAS data. Data analysis & annotation – New Bioconductor ready to read & query OWL/OBO into R. • Build in offline support for EFO & Bioportal ontology queries
② Onto. CAT & Zooma use cases 3. Local ontology management – e. Xtensive Genotype And Phenotype data platform (XGAP - Molgenis) : search widget Interactive annotation of data with ontology terms • 4. Allows search publically available ontologies & download terms for unambiguous annotation of QTL or GWAS data. Data analysis & annotation – New Bioconductor ready to read & query OWL/OBO into R. • Build in offline support for EFO & Bioportal ontology queries
 ② Onto. CAT characteristics & tools § Onto. CAT provides synonym & definition lookup across two major implemented ontology services § Supports interoperability using RDF § Class combining multiple ontology resources including different repositories behind single entry point (Composite. Ontology. Service) § Cache § Ranking Prioritization Fallback mechanism if ontology resource unavailable
② Onto. CAT characteristics & tools § Onto. CAT provides synonym & definition lookup across two major implemented ontology services § Supports interoperability using RDF § Class combining multiple ontology resources including different repositories behind single entry point (Composite. Ontology. Service) § Cache § Ranking Prioritization Fallback mechanism if ontology resource unavailable
 ②Demo on Google App Engine framework • http: //ontocat-web. appspot. com
②Demo on Google App Engine framework • http: //ontocat-web. appspot. com
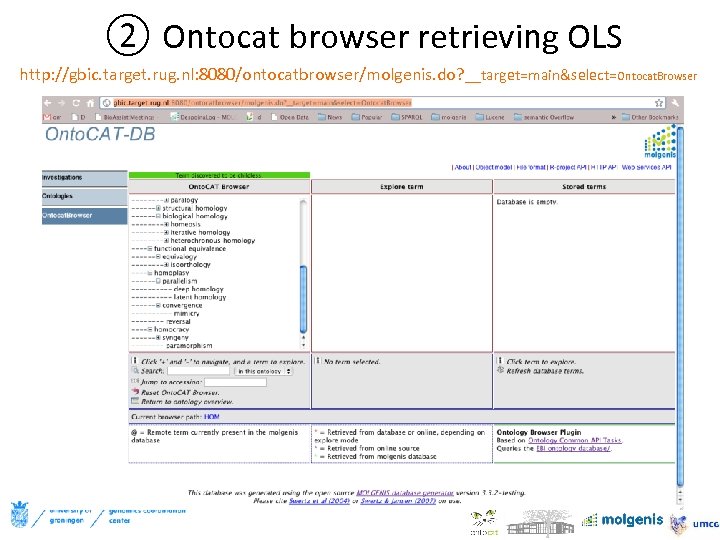 ② Ontocat browser retrieving OLS http: //gbic. target. rug. nl: 8080/ontocatbrowser/molgenis. do? __target=main&select=Ontocat. Browser
② Ontocat browser retrieving OLS http: //gbic. target. rug. nl: 8080/ontocatbrowser/molgenis. do? __target=main&select=Ontocat. Browser
 ②Onto. CAT’s applications • Onto. CAT ontology mapping application: – http: //zooma. sourceforge. net • Onto. CAT Bioconductor/R package: – http: //bioconductor. org/help/biocviews/2. 7/bioc/html/onto. CAT. html
②Onto. CAT’s applications • Onto. CAT ontology mapping application: – http: //zooma. sourceforge. net • Onto. CAT Bioconductor/R package: – http: //bioconductor. org/help/biocviews/2. 7/bioc/html/onto. CAT. html
 Outline • Three challenges for biologists’ and the corresponding for the Informatics’: 1. 2. 3. 4. Merge and pool data - Differences in formats and structure Find data - Differences in terminology Size of data - Automatic matching and merging Across data sets – All above + distribution • Approaches 1. 2. 3. 4. Integrate data into one ‘pheno’ model (MOLGENIS) Use ontologies (Onto. CAT) Indexing (Lucene) Query expansion (Lucene + Onto. CAT) • Discussion 1. Federated data queries (molgenis & rdf)
Outline • Three challenges for biologists’ and the corresponding for the Informatics’: 1. 2. 3. 4. Merge and pool data - Differences in formats and structure Find data - Differences in terminology Size of data - Automatic matching and merging Across data sets – All above + distribution • Approaches 1. 2. 3. 4. Integrate data into one ‘pheno’ model (MOLGENIS) Use ontologies (Onto. CAT) Indexing (Lucene) Query expansion (Lucene + Onto. CAT) • Discussion 1. Federated data queries (molgenis & rdf)
 ③Indexing: general features • • • Data structure overcomes barriers in large DB – created by using DB tables as basis for search – Efficient access of ordered records & rapid random lookup – Less disk space for storage (key fields) Open source java library (known in internet search engines) – Full text indexing & searching capability – Format independent (documents & fields) Query Expansion: – Add additional terms related (synonyms & children) appended by OR operator, assigned lower weight – Changes document ranking order of retrieved docs – Even if query expansion doesn’t improve search, query more precise DB
③Indexing: general features • • • Data structure overcomes barriers in large DB – created by using DB tables as basis for search – Efficient access of ordered records & rapid random lookup – Less disk space for storage (key fields) Open source java library (known in internet search engines) – Full text indexing & searching capability – Format independent (documents & fields) Query Expansion: – Add additional terms related (synonyms & children) appended by OR operator, assigned lower weight – Changes document ranking order of retrieved docs – Even if query expansion doesn’t improve search, query more precise DB
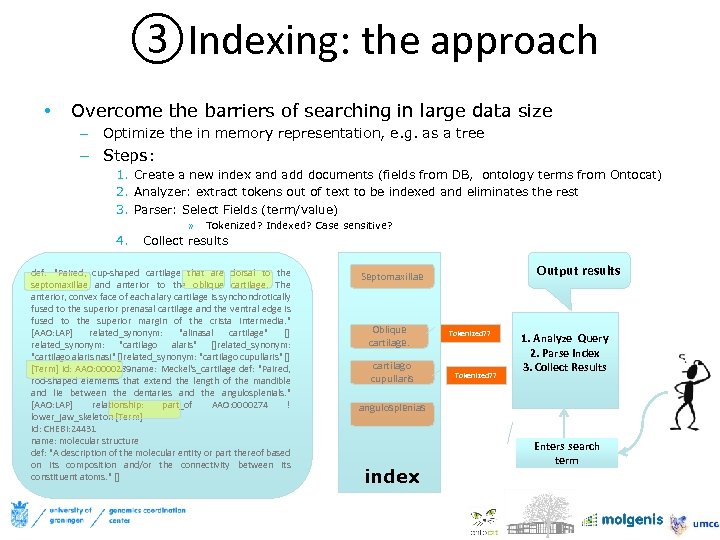 ③Indexing: the approach • Overcome the barriers of searching in large data size – Optimize the in memory representation, e. g. as a tree – Steps: 1. Create a new index and add documents (fields from DB, ontology terms from Ontocat) 2. Analyzer: extract tokens out of text to be indexed and eliminates the rest 3. Parser: Select Fields (term/value) » 4. Tokenized? Indexed? Case sensitive? Collect results def: "Paired, cup-shaped cartilage that are dorsal to the septomaxillae and anterior to the oblique cartilage. The anterior, convex face of each alary cartilage is synchondrotically fused to the superior prenasal cartilage and the ventral edge is fused to the superior margin of the crista intermedia. " [AAO: LAP] related_synonym: "alinasal cartilage" [] related_synonym: "cartilago alaris" []related_synonym: "cartilago alaris nasi" []related_synonym: "cartilago cupullaris" [] [Term] id: AAO: 0000289 name: Meckel's_cartilage def: "Paired, rod-shaped elements that extend the length of the mandible and lie between the dentaries and the angulosplenials. " [AAO: LAP] relationship: part_of AAO: 0000274 ! lower_jaw_skeleton [Term] id: CHEBI: 24431 name: molecular structure def: "A description of the molecular entity or part thereof based on its composition and/or the connectivity between its constituent atoms. " [] Output results Septomaxillae Oblique cartilage. cartilago cupullaris Tokenized? ? 1. Analyze Query 2. Parse Index 3. Collect Results angulosplenias index Enters search term
③Indexing: the approach • Overcome the barriers of searching in large data size – Optimize the in memory representation, e. g. as a tree – Steps: 1. Create a new index and add documents (fields from DB, ontology terms from Ontocat) 2. Analyzer: extract tokens out of text to be indexed and eliminates the rest 3. Parser: Select Fields (term/value) » 4. Tokenized? Indexed? Case sensitive? Collect results def: "Paired, cup-shaped cartilage that are dorsal to the septomaxillae and anterior to the oblique cartilage. The anterior, convex face of each alary cartilage is synchondrotically fused to the superior prenasal cartilage and the ventral edge is fused to the superior margin of the crista intermedia. " [AAO: LAP] related_synonym: "alinasal cartilage" [] related_synonym: "cartilago alaris" []related_synonym: "cartilago alaris nasi" []related_synonym: "cartilago cupullaris" [] [Term] id: AAO: 0000289 name: Meckel's_cartilage def: "Paired, rod-shaped elements that extend the length of the mandible and lie between the dentaries and the angulosplenials. " [AAO: LAP] relationship: part_of AAO: 0000274 ! lower_jaw_skeleton [Term] id: CHEBI: 24431 name: molecular structure def: "A description of the molecular entity or part thereof based on its composition and/or the connectivity between its constituent atoms. " [] Output results Septomaxillae Oblique cartilage. cartilago cupullaris Tokenized? ? 1. Analyze Query 2. Parse Index 3. Collect Results angulosplenias index Enters search term
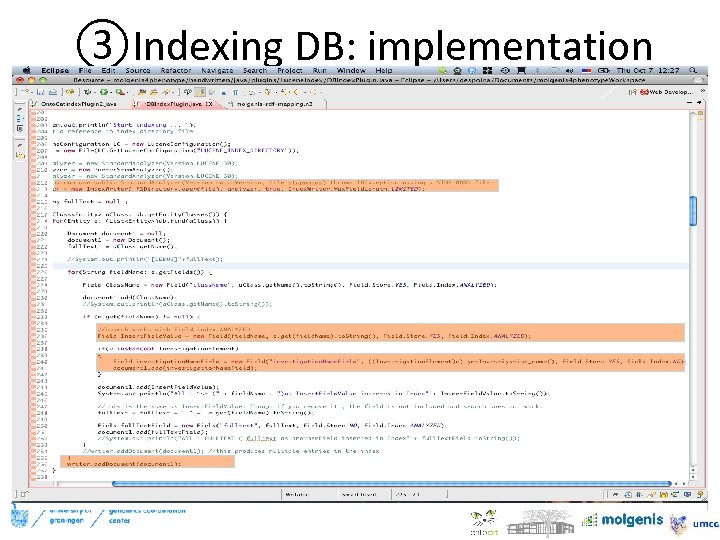 ③Indexing DB: implementation
③Indexing DB: implementation
 Outline • Three challenges for biologists’ and the corresponding for the Informatics’: 1. 2. 3. 4. Merge and pool data - Differences in formats and structure Find data - Differences in terminology Size of data - Automatic matching and merging Across data sets – All above + distribution • Approaches 1. 2. 3. 4. Integrate data into one ‘pheno’ model (MOLGENIS) Use ontologies (Onto. CAT) Indexing (Lucene) Query expansion (Lucene + Onto. CAT) • Discussion 1. Federated data queries (molgenis & rdf)
Outline • Three challenges for biologists’ and the corresponding for the Informatics’: 1. 2. 3. 4. Merge and pool data - Differences in formats and structure Find data - Differences in terminology Size of data - Automatic matching and merging Across data sets – All above + distribution • Approaches 1. 2. 3. 4. Integrate data into one ‘pheno’ model (MOLGENIS) Use ontologies (Onto. CAT) Indexing (Lucene) Query expansion (Lucene + Onto. CAT) • Discussion 1. Federated data queries (molgenis & rdf)
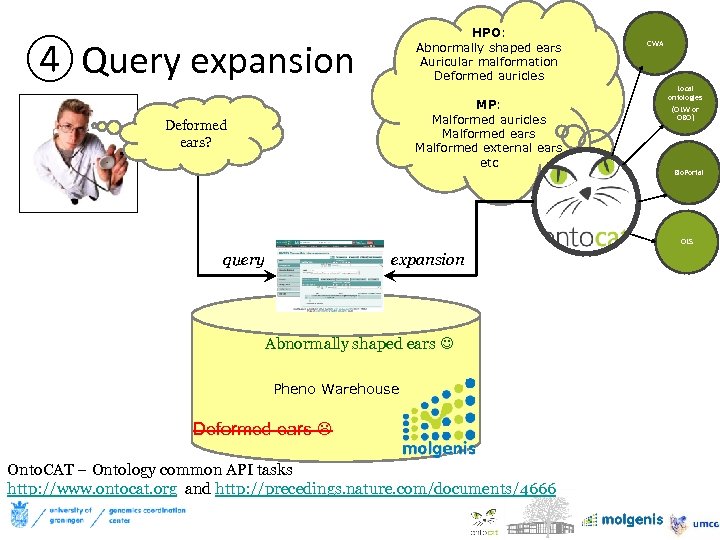 HPO: Abnormally shaped ears Auricular malformation Deformed auricles ④ Query expansion MP: Malformed auricles Malformed ears Malformed external ears etc Deformed ears? CWA 32 Local ontologies (OLW or OBO) Bio. Portal OLS query expansion Abnormally shaped ears Pheno Warehouse Deformed ears Onto. CAT – Ontology common API tasks http: //www. ontocat. org and http: //precedings. nature. com/documents/4666
HPO: Abnormally shaped ears Auricular malformation Deformed auricles ④ Query expansion MP: Malformed auricles Malformed ears Malformed external ears etc Deformed ears? CWA 32 Local ontologies (OLW or OBO) Bio. Portal OLS query expansion Abnormally shaped ears Pheno Warehouse Deformed ears Onto. CAT – Ontology common API tasks http: //www. ontocat. org and http: //precedings. nature. com/documents/4666
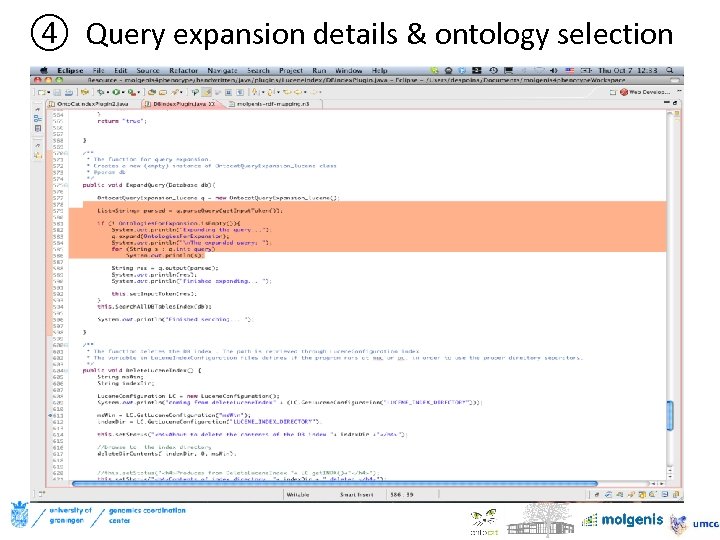 ④ Query expansion details & ontology selection Ontologies used
④ Query expansion details & ontology selection Ontologies used
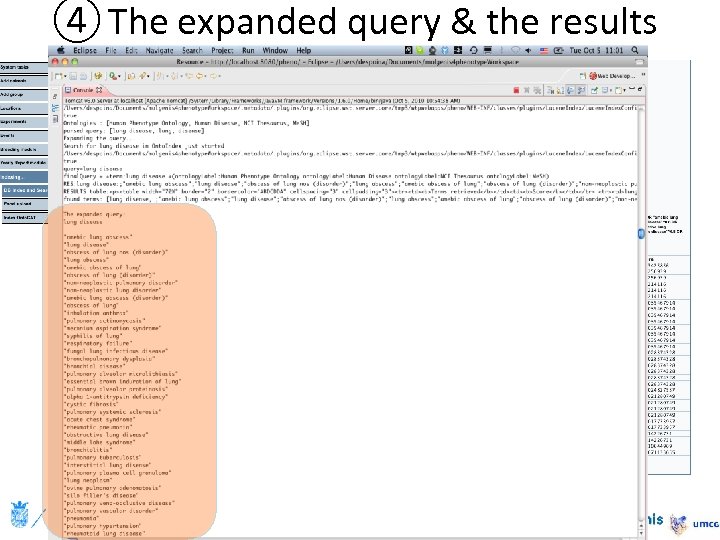 ④ The expanded query & the results
④ The expanded query & the results
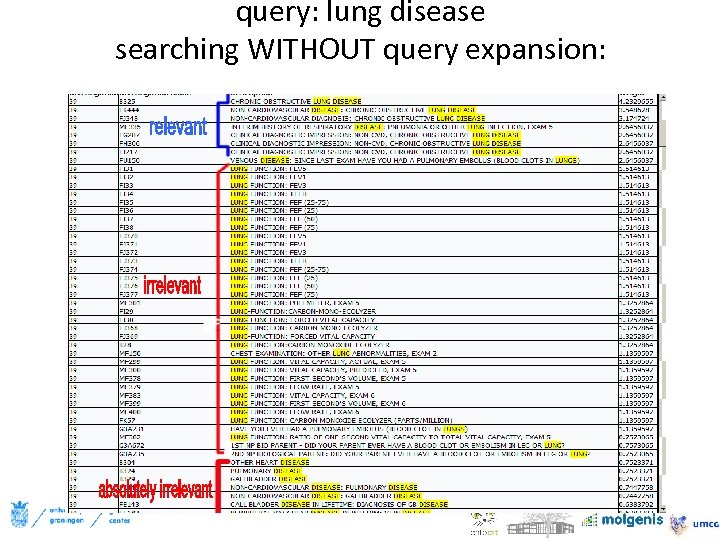 query: lung disease searching WITHOUT query expansion:
query: lung disease searching WITHOUT query expansion:
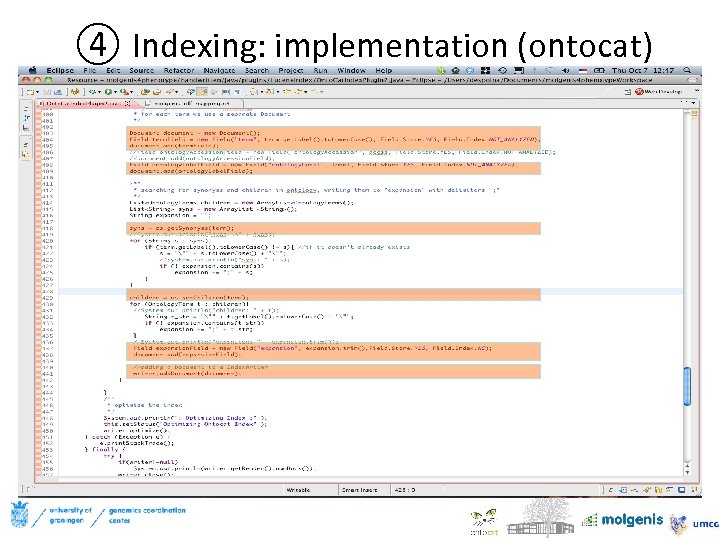 ④ Indexing: implementation (ontocat) Lucene scoring uses a combination of the Vector Space Model (VSM) of Information Retrieval and the Boolean model to determine how relevant a given Document is to a User's query.
④ Indexing: implementation (ontocat) Lucene scoring uses a combination of the Vector Space Model (VSM) of Information Retrieval and the Boolean model to determine how relevant a given Document is to a User's query.
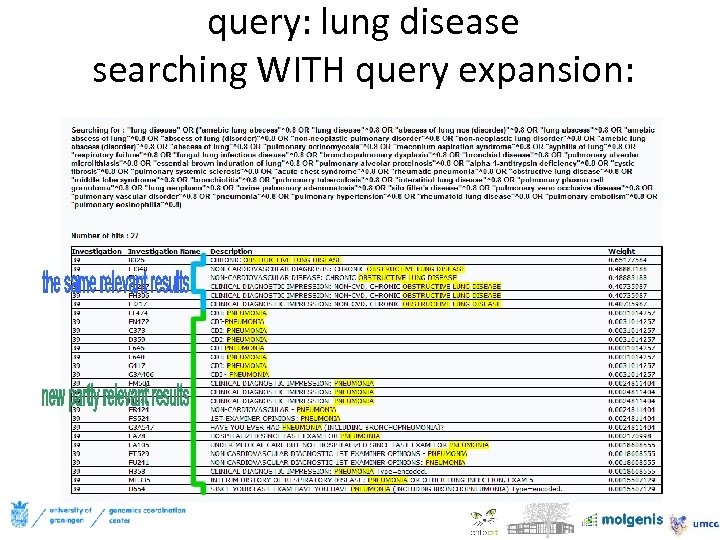 query: lung disease searching WITH query expansion:
query: lung disease searching WITH query expansion:
 Outline • Three challenges for biologists’ and the corresponding for the Informatics’: 1. 2. 3. 4. Merge and pool data - Differences in formats and structure Find data - Differences in terminology Size of data - Automatic matching and merging Across data sets – All above + distribution • Approaches 1. 2. 3. 4. Integrate data into one ‘pheno’ model (MOLGENIS) Use ontologies (Onto. CAT) Indexing (Lucene) Query expansion (Lucene + Onto. CAT) • Discussion 1. Federated data queries (molgenis & rdf)
Outline • Three challenges for biologists’ and the corresponding for the Informatics’: 1. 2. 3. 4. Merge and pool data - Differences in formats and structure Find data - Differences in terminology Size of data - Automatic matching and merging Across data sets – All above + distribution • Approaches 1. 2. 3. 4. Integrate data into one ‘pheno’ model (MOLGENIS) Use ontologies (Onto. CAT) Indexing (Lucene) Query expansion (Lucene + Onto. CAT) • Discussion 1. Federated data queries (molgenis & rdf)
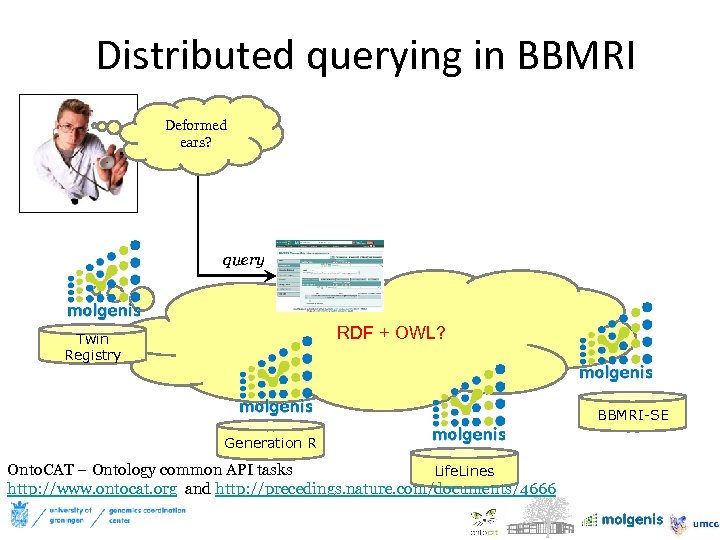 Distributed querying in BBMRI Deformed ears? query RDF + OWL? Twin Registry BBMRI-SE Generation R Onto. CAT – Ontology common API tasks Life. Lines http: //www. ontocat. org and http: //precedings. nature. com/documents/4666
Distributed querying in BBMRI Deformed ears? query RDF + OWL? Twin Registry BBMRI-SE Generation R Onto. CAT – Ontology common API tasks Life. Lines http: //www. ontocat. org and http: //precedings. nature. com/documents/4666
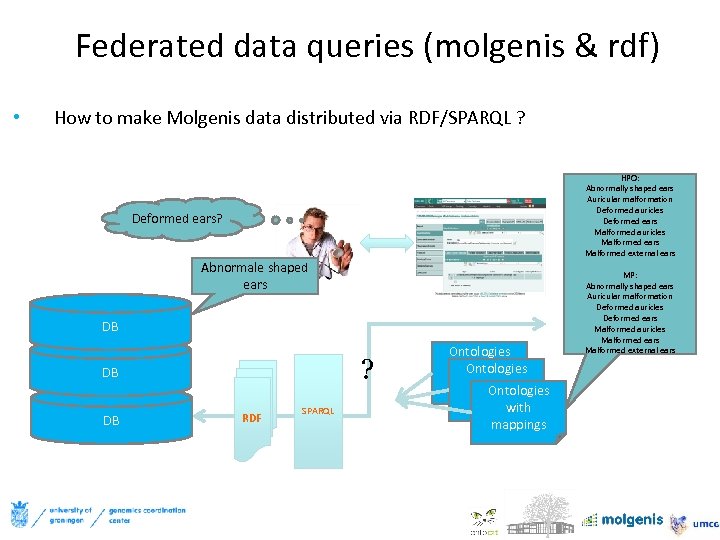 Federated data queries (molgenis & rdf) • How to make Molgenis data distributed via RDF/SPARQL ? HPO: Abnormally shaped ears Auricular malformation Deformed auricles Deformed ears Malformed auricles Malformed ears Malformed external ears Deformed ears? Abnormale shaped ears DB ? DB DB RDF SPARQL Ontologies with Ontologies mappings with mappings MP: Abnormally shaped ears Auricular malformation Deformed auricles Deformed ears Malformed auricles Malformed ears Malformed external ears
Federated data queries (molgenis & rdf) • How to make Molgenis data distributed via RDF/SPARQL ? HPO: Abnormally shaped ears Auricular malformation Deformed auricles Deformed ears Malformed auricles Malformed ears Malformed external ears Deformed ears? Abnormale shaped ears DB ? DB DB RDF SPARQL Ontologies with Ontologies mappings with mappings MP: Abnormally shaped ears Auricular malformation Deformed auricles Deformed ears Malformed auricles Malformed ears Malformed external ears
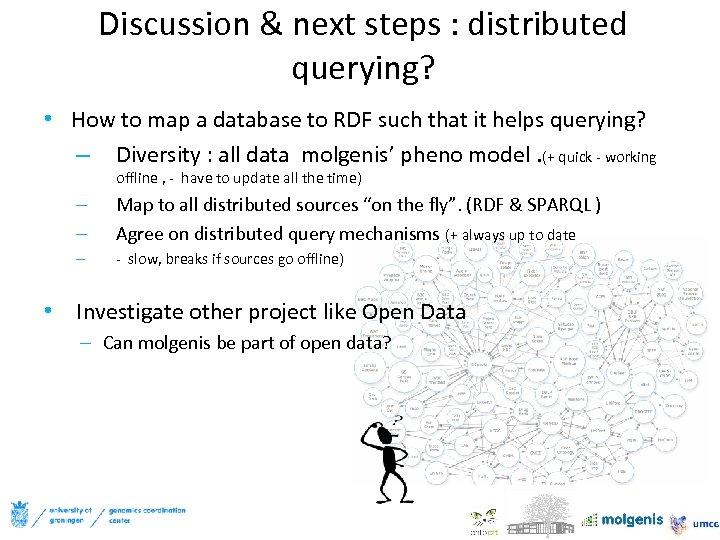 Discussion & next steps : distributed querying? • How to map a database to RDF such that it helps querying? – Diversity : all data molgenis’ pheno model. (+ quick - working offline , - have to update all the time) – – Map to all distributed sources “on the fly”. (RDF & SPARQL ) Agree on distributed query mechanisms (+ always up to date – - slow, breaks if sources go offline) • Investigate other project like Open Data – Can molgenis be part of open data?
Discussion & next steps : distributed querying? • How to map a database to RDF such that it helps querying? – Diversity : all data molgenis’ pheno model. (+ quick - working offline , - have to update all the time) – – Map to all distributed sources “on the fly”. (RDF & SPARQL ) Agree on distributed query mechanisms (+ always up to date – - slow, breaks if sources go offline) • Investigate other project like Open Data – Can molgenis be part of open data?
 NL NL
NL NL
 Thank you for your attention. Questions?
Thank you for your attention. Questions?
 • Ontocat http: //www. ontocat. org/ , – http: //precedings. nature. com/documents/4666/version/1 – http: //www. biomedcentral. com/imedia/1627447285460829_article. pdf – Guide/ examples http: //www. ontocat. org/wiki/Ontocat. Guide – Available from : – http: //gbic. target. rug. nl: 8080/ontocatbrowser/molgenis. do? __targe t=main&select=Ontocat. Browser – Ontocat Demo on Google App Engine framework : http: //ontocatweb. appspot. com • Molgenis Lucene Index & query expansion app : – http: //www. molgenis. org/svn/molgenis_projects/molgenis 4 phenot ype/handwritten/java/plugins/Lucene. Index/ • Pheno-OM datamodel : http: //wwwdev. ebi. ac. uk/microarraysrv/pheno/doc/objectmodel. html • XGAP: http: //www. xgap. org
• Ontocat http: //www. ontocat. org/ , – http: //precedings. nature. com/documents/4666/version/1 – http: //www. biomedcentral. com/imedia/1627447285460829_article. pdf – Guide/ examples http: //www. ontocat. org/wiki/Ontocat. Guide – Available from : – http: //gbic. target. rug. nl: 8080/ontocatbrowser/molgenis. do? __targe t=main&select=Ontocat. Browser – Ontocat Demo on Google App Engine framework : http: //ontocatweb. appspot. com • Molgenis Lucene Index & query expansion app : – http: //www. molgenis. org/svn/molgenis_projects/molgenis 4 phenot ype/handwritten/java/plugins/Lucene. Index/ • Pheno-OM datamodel : http: //wwwdev. ebi. ac. uk/microarraysrv/pheno/doc/objectmodel. html • XGAP: http: //www. xgap. org


