2a3193e574a80e1538e420cf430e5f8f.ppt
- Количество слайдов: 1
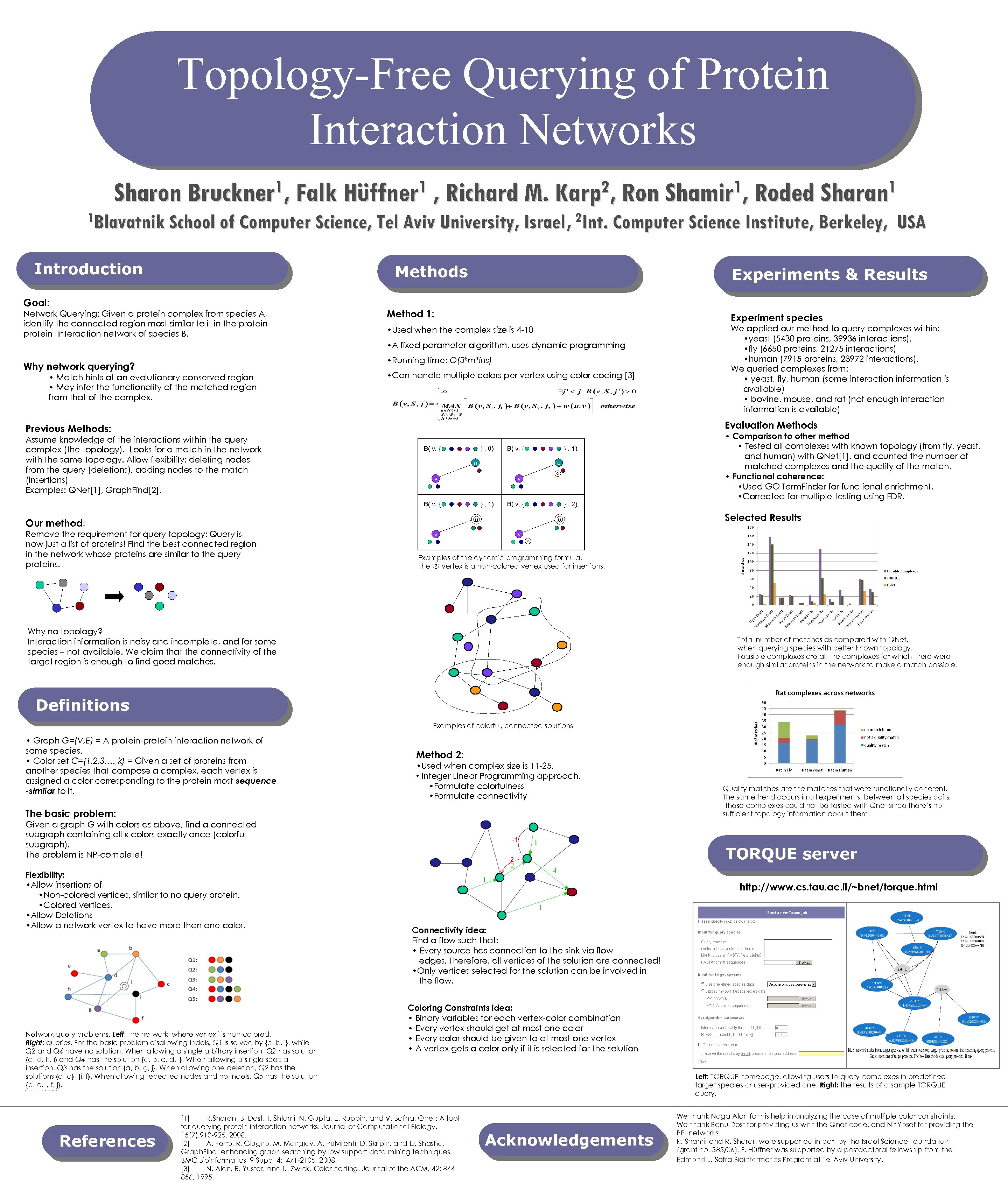 Topology-Free Querying of Protein Interaction Networks 1, Falk Hüffner 1 , Richard M. Karp 2, Ron Shamir 1, Roded Sharan 1 Sharon Bruckner 1 Blavatnik School of Computer Science, Tel Aviv University, Israel , 2 Int. Computer Science Institute, Berkeley, Introduction Methods Goal: Network Querying: Given a protein complex from species A, identify the connected region most similar to it in the protein Interaction network of species B. Experiments & Results Method 1: Experiment species • Used when the complex size is 4 -10 We applied our method to query complexes within: • yeast (5430 proteins, 39936 interactions), • fly (6650 proteins, 21275 interactions) • human (7915 proteins, 28972 interactions). We queried complexes from: • yeast, fly, human (some interaction information is available) • bovine, mouse, and rat (not enough interaction information is available) • A fixed parameter algorithm, uses dynamic programming • Running time: O(3 km*ins) Why network querying? • Match hints at an evolutionary conserved region • May infer the functionality of the matched region from that of the complex. USA • Can handle multiple colors per vertex using color coding [3] Evaluation Methods Previous Methods: Assume knowledge of the interactions within the query complex (the topology). Looks for a match in the network with the same topology. Allow flexibility: deleting nodes from the query (deletions), adding nodes to the match (insertions) Examples: QNet[1], Graph. Find[2]. B( v, { } , 0) B( v, { u u v v B( v, { } , 1) B( v, { u Our method: Remove the requirement for query topology: Query is now just a list of proteins! Find the best connected region in the network whose proteins are similar to the query proteins. } , 1) } , 2) u v • Comparison to other method • Tested all complexes with known topology (from fly, yeast, and human) with QNet[1], and counted the number of matched complexes and the quality of the match. • Functional coherence: • Used GO Term. Finder for functional enrichment. • Corrected for multiple testing using FDR. Selected Results v Examples of the dynamic programming formula. The vertex is a non-colored vertex used for insertions. Why no topology? Interaction information is noisy and incomplete, and for some species – not available. We claim that the connectivity of the target region is enough to find good matches. Total number of matches as compared with QNet, when querying species with better known topology. Feasible complexes are all the complexes for which there were enough similar proteins in the network to make a match possible. Definitions Examples of colorful, connected solutions • Graph G=(V, E) = A protein-protein interaction network of some species. • Color set C={1, 2, 3, …, k} = Given a set of proteins from another species that compose a complex, each vertex is assigned a color corresponding to the protein most sequence -similar to it. Method 2: • Used when complex size is 11 -25. • Integer Linear Programming approach. • Formulate colorfulness • Formulate connectivity The basic problem: Given a graph G with colors as above, find a connected subgraph containing all k colors exactly once (colorful subgraph). The problem is NP-complete! Flexibility: • Allow insertions of • Non-colored vertices, similar to no query protein. • Colored vertices. • Allow Deletions • Allow a network vertex to have more than one color. Network query problems. Left: the network, where vertex j is non-colored. Right: queries. For the basic problem disallowing indels, Q 1 is solved by {c, b, i}, while Q 2 and Q 4 have no solution. When allowing a single arbitrary insertion, Q 2 has solution {a, d, h, i} and Q 4 has the solution {a, b, c, d, i}. When allowing a single special insertion, Q 3 has the solution {a, b, g, j}. When allowing one deletion, Q 2 has the solutions {a, d}, {i, f}. When allowing repeated nodes and no indels, Q 5 has the solution {b, c, I, f, j}. References Quality matches are the matches that were functionally coherent. The same trend occurs in all experiments, between all species pairs. These complexes could not be tested with Qnet since there’s no sufficient topology information about them. -1 -2 TORQUE server http: //www. cs. tau. ac. il/~bnet/torque. html Connectivity idea: Find a flow such that: • Every source has connection to the sink via flow edges. Therefore, all vertices of the solution are connected! • Only vertices selected for the solution can be involved in the flow. Coloring Constraints idea: • Binary variables for each vertex-color combination • Every vertex should get at most one color • Every color should be given to at most one vertex • A vertex gets a color only if it is selected for the solution [1] R. Sharan, B. Dost, T. Shlomi, N. Gupta, E. Ruppin, and V. Bafna. Qnet: A tool for querying protein interaction networks. Journal of Computational Biology, 15(7): 913 -925, 2008. [2] A. Ferro, R. Giugno, M. Mongiov, A. Pulvirenti, D. Skripin, and D. Shasha. Graph. Find: enhancing graph searching by low support data mining techniques. BMC Bioinformatics, 9 Suppl 4: 1471 -2105, 2008. [3] N. Alon, R. Yuster, and U. Zwick. Color coding. Journal of the ACM, 42: 844856, 1995. Left: TORQUE homepage, allowing users to query complexes in predefined target species or user-provided one. Right: the results of a sample TORQUE query. Acknowledgements We thank Noga Alon for his help in analyzing the case of multiple color constraints. We thank Banu Dost for providing us with the Qnet code, and Nir Yosef for providing the PPI networks. R. Shamir and R. Sharan were supported in part by the Israel Science Foundation (grant no. 385/06). F. Hüffner was supported by a postdoctoral fellowship from the Edmond J. Safra Bioinformatics Program at Tel Aviv University.
Topology-Free Querying of Protein Interaction Networks 1, Falk Hüffner 1 , Richard M. Karp 2, Ron Shamir 1, Roded Sharan 1 Sharon Bruckner 1 Blavatnik School of Computer Science, Tel Aviv University, Israel , 2 Int. Computer Science Institute, Berkeley, Introduction Methods Goal: Network Querying: Given a protein complex from species A, identify the connected region most similar to it in the protein Interaction network of species B. Experiments & Results Method 1: Experiment species • Used when the complex size is 4 -10 We applied our method to query complexes within: • yeast (5430 proteins, 39936 interactions), • fly (6650 proteins, 21275 interactions) • human (7915 proteins, 28972 interactions). We queried complexes from: • yeast, fly, human (some interaction information is available) • bovine, mouse, and rat (not enough interaction information is available) • A fixed parameter algorithm, uses dynamic programming • Running time: O(3 km*ins) Why network querying? • Match hints at an evolutionary conserved region • May infer the functionality of the matched region from that of the complex. USA • Can handle multiple colors per vertex using color coding [3] Evaluation Methods Previous Methods: Assume knowledge of the interactions within the query complex (the topology). Looks for a match in the network with the same topology. Allow flexibility: deleting nodes from the query (deletions), adding nodes to the match (insertions) Examples: QNet[1], Graph. Find[2]. B( v, { } , 0) B( v, { u u v v B( v, { } , 1) B( v, { u Our method: Remove the requirement for query topology: Query is now just a list of proteins! Find the best connected region in the network whose proteins are similar to the query proteins. } , 1) } , 2) u v • Comparison to other method • Tested all complexes with known topology (from fly, yeast, and human) with QNet[1], and counted the number of matched complexes and the quality of the match. • Functional coherence: • Used GO Term. Finder for functional enrichment. • Corrected for multiple testing using FDR. Selected Results v Examples of the dynamic programming formula. The vertex is a non-colored vertex used for insertions. Why no topology? Interaction information is noisy and incomplete, and for some species – not available. We claim that the connectivity of the target region is enough to find good matches. Total number of matches as compared with QNet, when querying species with better known topology. Feasible complexes are all the complexes for which there were enough similar proteins in the network to make a match possible. Definitions Examples of colorful, connected solutions • Graph G=(V, E) = A protein-protein interaction network of some species. • Color set C={1, 2, 3, …, k} = Given a set of proteins from another species that compose a complex, each vertex is assigned a color corresponding to the protein most sequence -similar to it. Method 2: • Used when complex size is 11 -25. • Integer Linear Programming approach. • Formulate colorfulness • Formulate connectivity The basic problem: Given a graph G with colors as above, find a connected subgraph containing all k colors exactly once (colorful subgraph). The problem is NP-complete! Flexibility: • Allow insertions of • Non-colored vertices, similar to no query protein. • Colored vertices. • Allow Deletions • Allow a network vertex to have more than one color. Network query problems. Left: the network, where vertex j is non-colored. Right: queries. For the basic problem disallowing indels, Q 1 is solved by {c, b, i}, while Q 2 and Q 4 have no solution. When allowing a single arbitrary insertion, Q 2 has solution {a, d, h, i} and Q 4 has the solution {a, b, c, d, i}. When allowing a single special insertion, Q 3 has the solution {a, b, g, j}. When allowing one deletion, Q 2 has the solutions {a, d}, {i, f}. When allowing repeated nodes and no indels, Q 5 has the solution {b, c, I, f, j}. References Quality matches are the matches that were functionally coherent. The same trend occurs in all experiments, between all species pairs. These complexes could not be tested with Qnet since there’s no sufficient topology information about them. -1 -2 TORQUE server http: //www. cs. tau. ac. il/~bnet/torque. html Connectivity idea: Find a flow such that: • Every source has connection to the sink via flow edges. Therefore, all vertices of the solution are connected! • Only vertices selected for the solution can be involved in the flow. Coloring Constraints idea: • Binary variables for each vertex-color combination • Every vertex should get at most one color • Every color should be given to at most one vertex • A vertex gets a color only if it is selected for the solution [1] R. Sharan, B. Dost, T. Shlomi, N. Gupta, E. Ruppin, and V. Bafna. Qnet: A tool for querying protein interaction networks. Journal of Computational Biology, 15(7): 913 -925, 2008. [2] A. Ferro, R. Giugno, M. Mongiov, A. Pulvirenti, D. Skripin, and D. Shasha. Graph. Find: enhancing graph searching by low support data mining techniques. BMC Bioinformatics, 9 Suppl 4: 1471 -2105, 2008. [3] N. Alon, R. Yuster, and U. Zwick. Color coding. Journal of the ACM, 42: 844856, 1995. Left: TORQUE homepage, allowing users to query complexes in predefined target species or user-provided one. Right: the results of a sample TORQUE query. Acknowledgements We thank Noga Alon for his help in analyzing the case of multiple color constraints. We thank Banu Dost for providing us with the Qnet code, and Nir Yosef for providing the PPI networks. R. Shamir and R. Sharan were supported in part by the Israel Science Foundation (grant no. 385/06). F. Hüffner was supported by a postdoctoral fellowship from the Edmond J. Safra Bioinformatics Program at Tel Aviv University.


