6e8245d20bf5a870aae1e2854814ebff.ppt
- Количество слайдов: 59
 This Week • Chapters 9, 10. 1 and 10. 4 - 10. 7 for reference, – exam material will be on lecture content for the above, – sample questions, and questions for these chapters will be posted this afternoon. • Exam Friday: Assignments in Chapters 6, 11, 8, 9 and 10. All lecture material.
This Week • Chapters 9, 10. 1 and 10. 4 - 10. 7 for reference, – exam material will be on lecture content for the above, – sample questions, and questions for these chapters will be posted this afternoon. • Exam Friday: Assignments in Chapters 6, 11, 8, 9 and 10. All lecture material.
 Gene Expression …the processes by which information contained in genes and genomes is decoded by cells, . . . in order to produce molecules that determine the phenotypes observed in organisms, – transcription (post-transcriptional modifications), – translation (post-translational modifications.
Gene Expression …the processes by which information contained in genes and genomes is decoded by cells, . . . in order to produce molecules that determine the phenotypes observed in organisms, – transcription (post-transcriptional modifications), – translation (post-translational modifications.
 Transcription. . . the synthesis of m. RNA from a DNA template, - now it is important to understand when and where, as well as how.
Transcription. . . the synthesis of m. RNA from a DNA template, - now it is important to understand when and where, as well as how.
 m. RNA Synthesis • Template (DNA) and Promoter, • Nucleoside triphosphates (NTPs), – N: A, U, G, or C, • Enzymes (RNA polymerases), • Energy (as in replication, from phosphate bonds).
m. RNA Synthesis • Template (DNA) and Promoter, • Nucleoside triphosphates (NTPs), – N: A, U, G, or C, • Enzymes (RNA polymerases), • Energy (as in replication, from phosphate bonds).
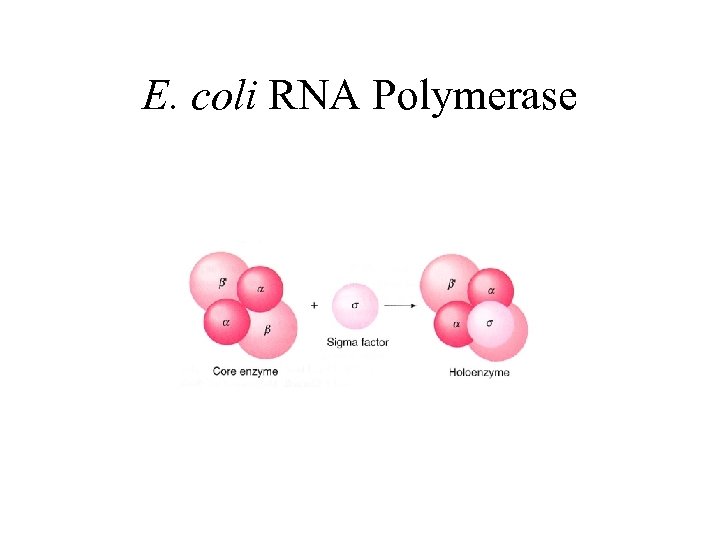 E. coli RNA Polymerase
E. coli RNA Polymerase
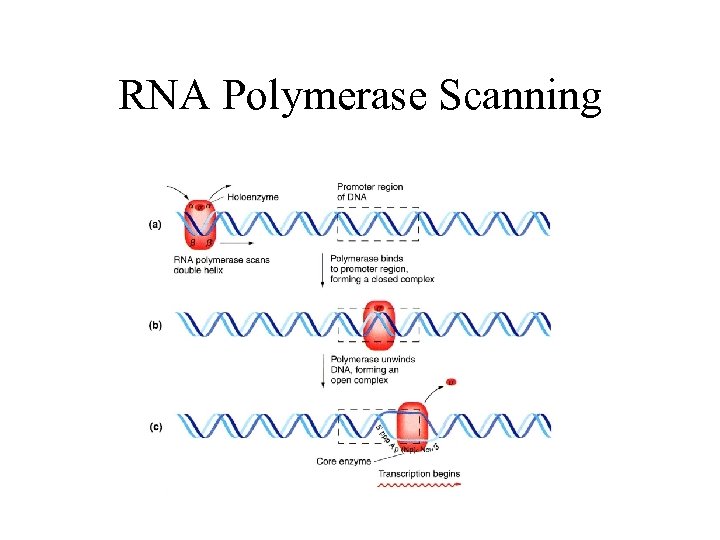 RNA Polymerase Scanning
RNA Polymerase Scanning
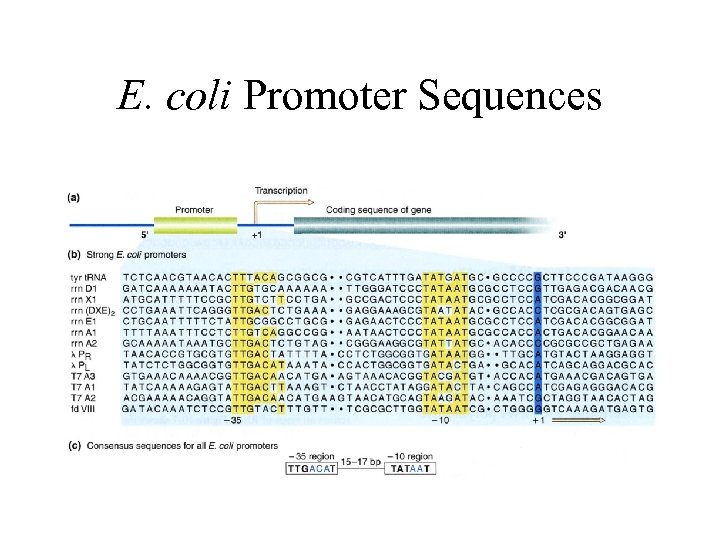 E. coli Promoter Sequences
E. coli Promoter Sequences
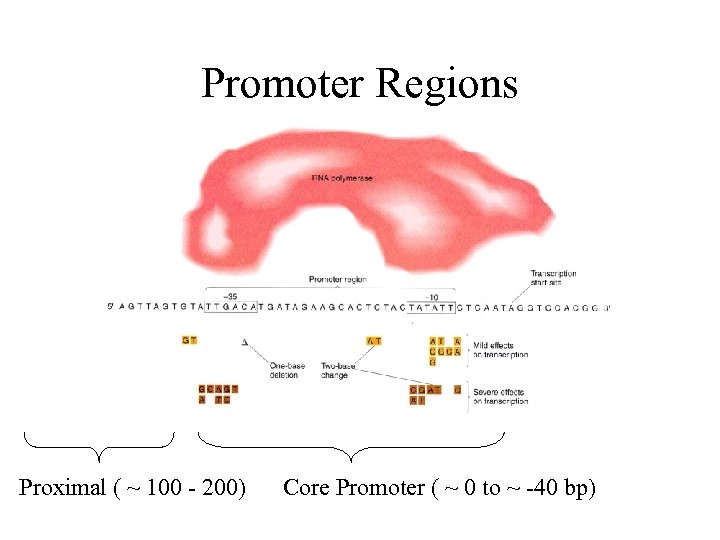 Promoter Regions Proximal ( ~ 100 - 200) Core Promoter ( ~ 0 to ~ -40 bp)
Promoter Regions Proximal ( ~ 100 - 200) Core Promoter ( ~ 0 to ~ -40 bp)
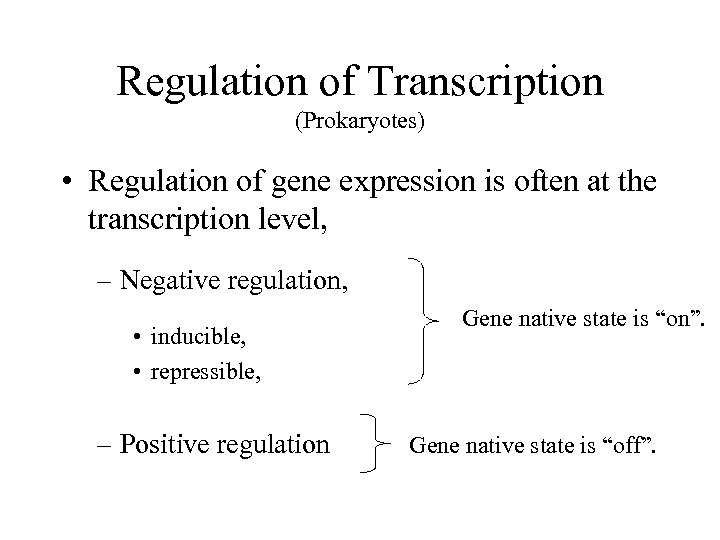 Regulation of Transcription (Prokaryotes) • Regulation of gene expression is often at the transcription level, – Negative regulation, • inducible, • repressible, – Positive regulation Gene native state is “on”. Gene native state is “off”.
Regulation of Transcription (Prokaryotes) • Regulation of gene expression is often at the transcription level, – Negative regulation, • inducible, • repressible, – Positive regulation Gene native state is “on”. Gene native state is “off”.
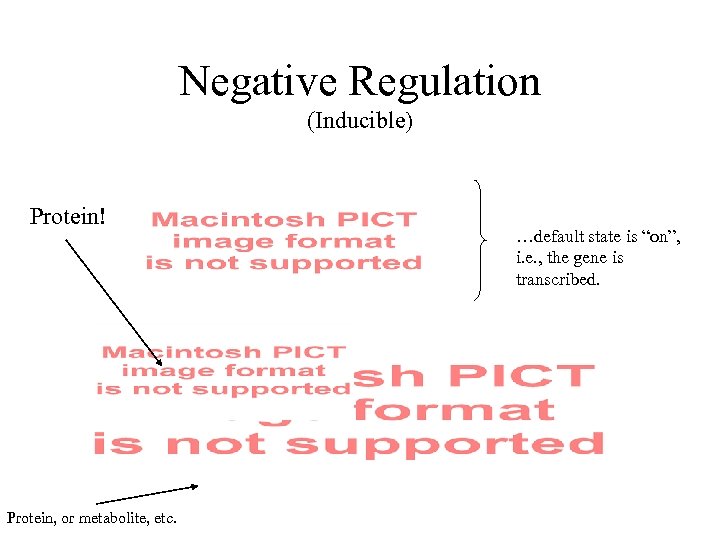 Negative Regulation (Inducible) Protein! Protein, or metabolite, etc. …default state is “on”, i. e. , the gene is transcribed.
Negative Regulation (Inducible) Protein! Protein, or metabolite, etc. …default state is “on”, i. e. , the gene is transcribed.
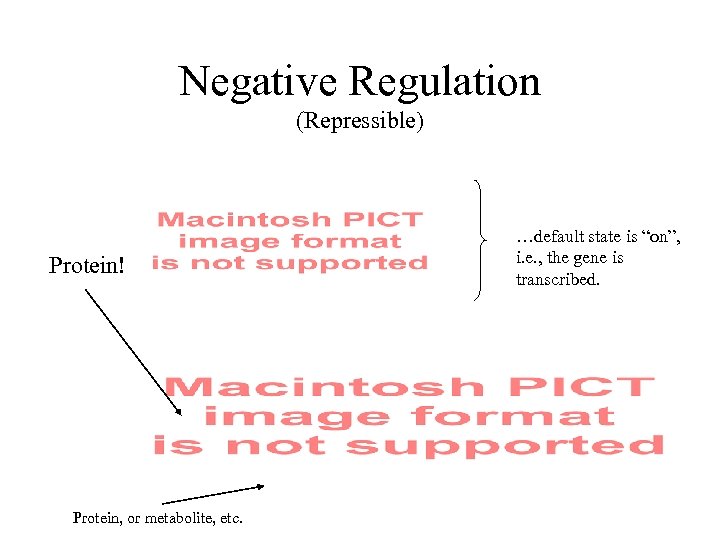 Negative Regulation (Repressible) Protein! Protein, or metabolite, etc. …default state is “on”, i. e. , the gene is transcribed.
Negative Regulation (Repressible) Protein! Protein, or metabolite, etc. …default state is “on”, i. e. , the gene is transcribed.
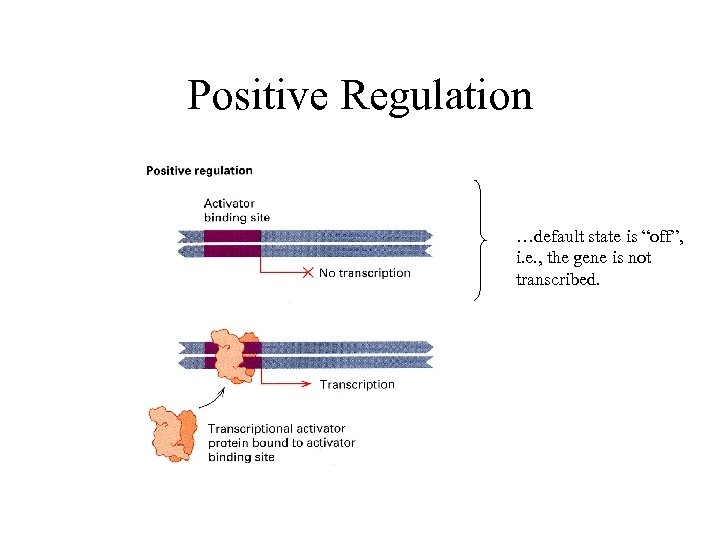 Positive Regulation …default state is “off”, i. e. , the gene is not transcribed.
Positive Regulation …default state is “off”, i. e. , the gene is not transcribed.
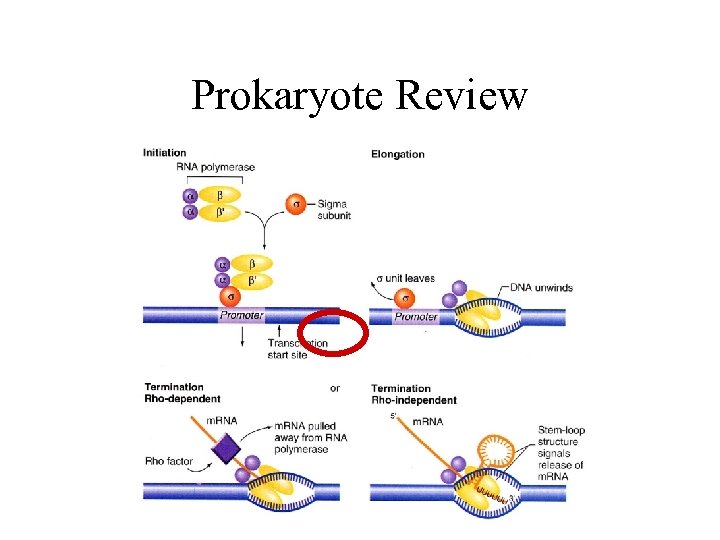 Prokaryote Review
Prokaryote Review
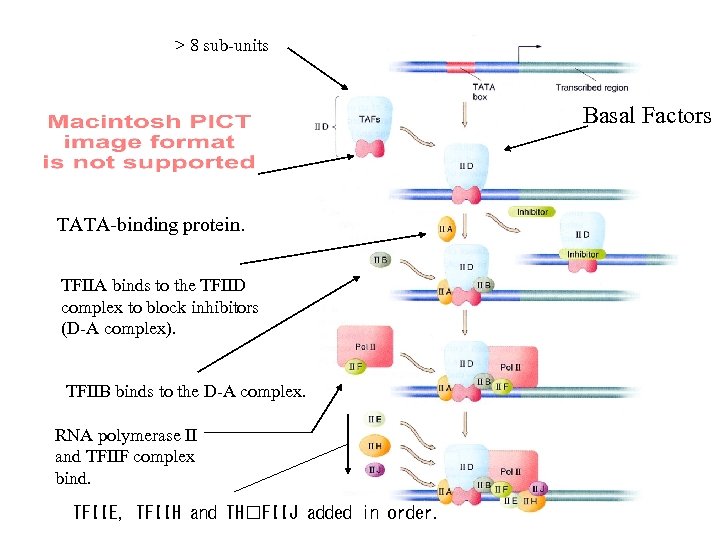 > 8 sub-units Basal Factors TATA-binding protein. TFIIA binds to the TFIID complex to block inhibitors (D-A complex). TFIIB binds to the D-A complex. RNA polymerase II and TFIIF complex bind. TFIIE, TFIIH and TH FIIJ added in order.
> 8 sub-units Basal Factors TATA-binding protein. TFIIA binds to the TFIID complex to block inhibitors (D-A complex). TFIIB binds to the D-A complex. RNA polymerase II and TFIIF complex bind. TFIIE, TFIIH and TH FIIJ added in order.
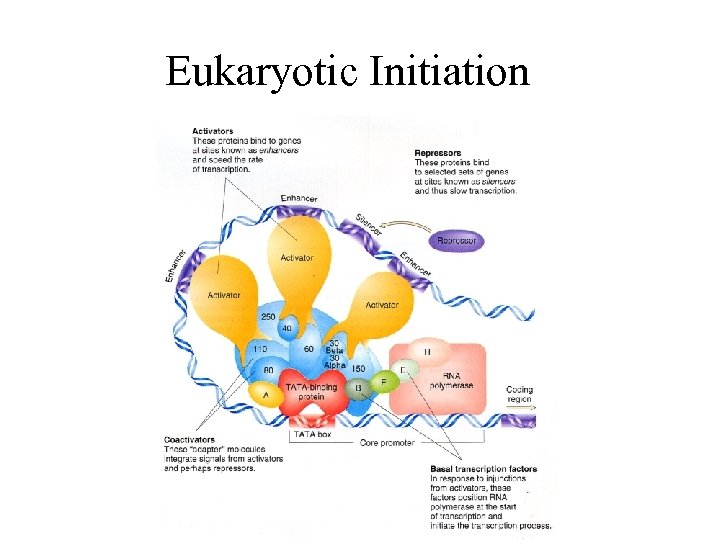 Eukaryotic Initiation
Eukaryotic Initiation
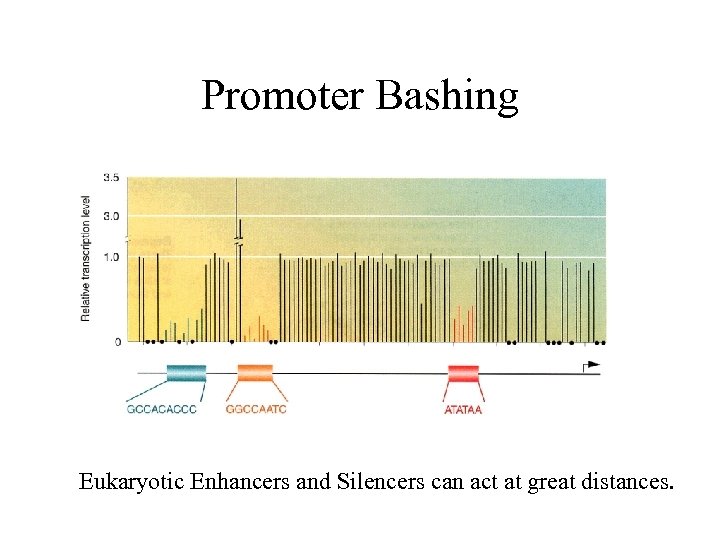 Promoter Bashing Eukaryotic Enhancers and Silencers can act at great distances.
Promoter Bashing Eukaryotic Enhancers and Silencers can act at great distances.
 Drosophila Guts and Such dpp locus . . . (c) and (d) are ID enhancer driven, give rise to fly appenages.
Drosophila Guts and Such dpp locus . . . (c) and (d) are ID enhancer driven, give rise to fly appenages.
 Terms • cis-acting elements; – DNA sequences that serve as attachments sites for the DNAbinding proteins that regulate the initiation of transcription. • trans-acting elements; – the DNA-binding proteins that regulate the initiation of transcription.
Terms • cis-acting elements; – DNA sequences that serve as attachments sites for the DNAbinding proteins that regulate the initiation of transcription. • trans-acting elements; – the DNA-binding proteins that regulate the initiation of transcription.
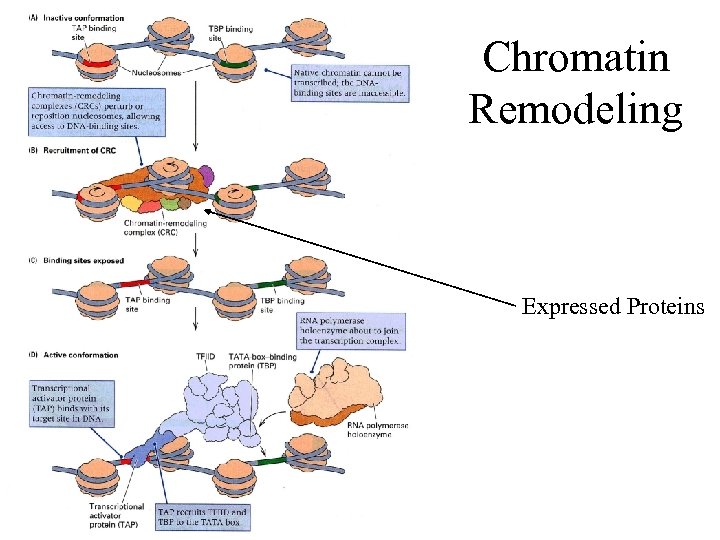 Chromatin Remodeling Expressed Proteins
Chromatin Remodeling Expressed Proteins
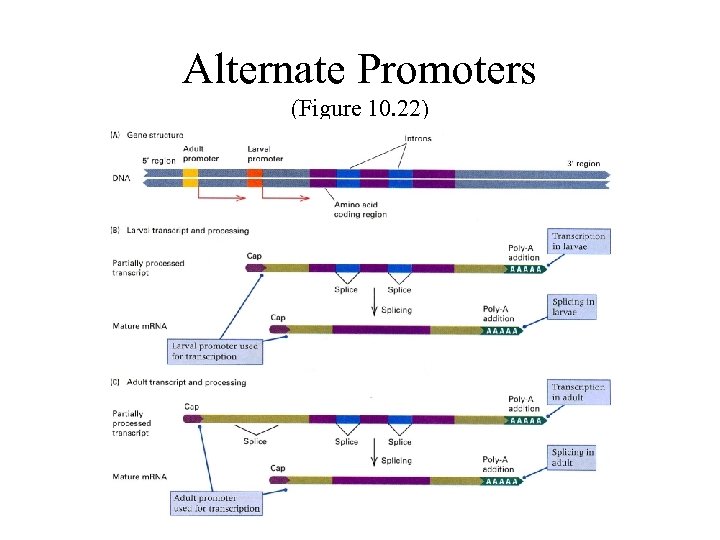 Alternate Promoters (Figure 10. 22)
Alternate Promoters (Figure 10. 22)
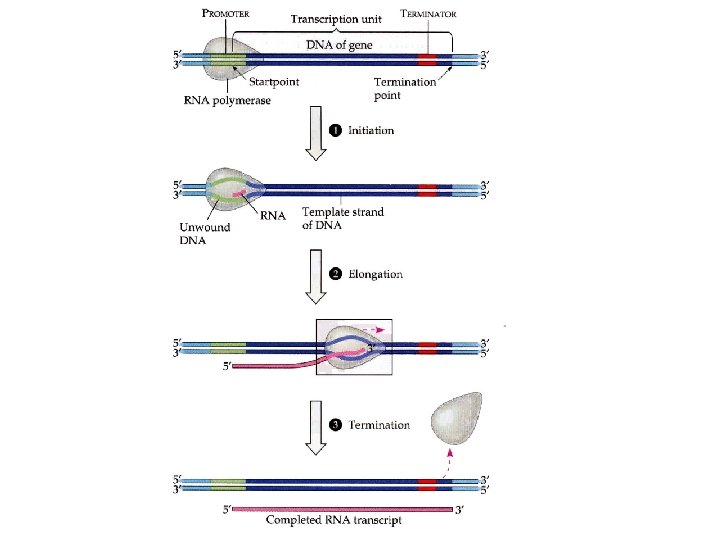
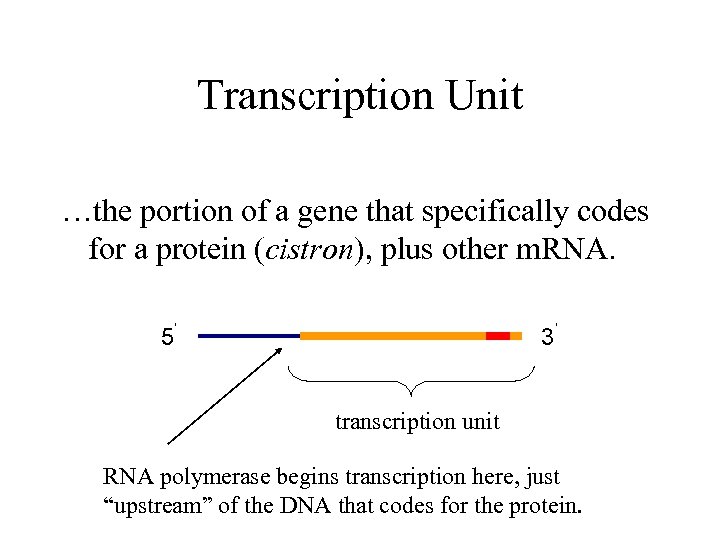 Transcription Unit …the portion of a gene that specifically codes for a protein (cistron), plus other m. RNA. 5’ 3’ transcription unit RNA polymerase begins transcription here, just “upstream” of the DNA that codes for the protein.
Transcription Unit …the portion of a gene that specifically codes for a protein (cistron), plus other m. RNA. 5’ 3’ transcription unit RNA polymerase begins transcription here, just “upstream” of the DNA that codes for the protein.
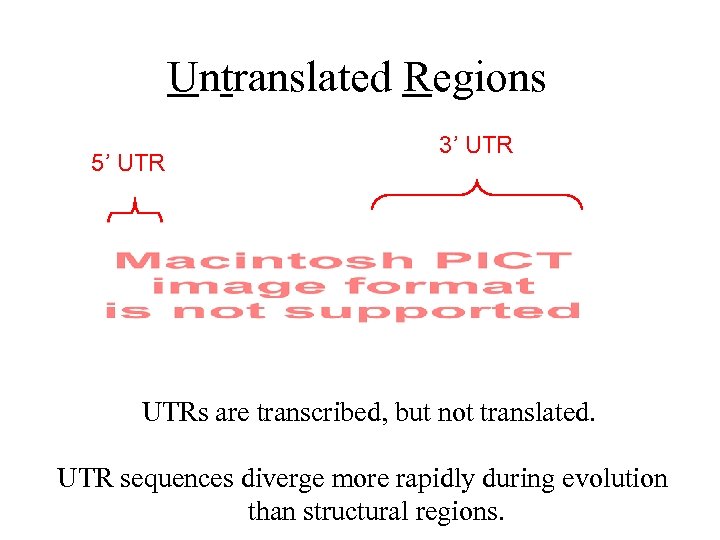 Untranslated Regions 5’ UTR 3’ UTRs are transcribed, but not translated. UTR sequences diverge more rapidly during evolution than structural regions.
Untranslated Regions 5’ UTR 3’ UTRs are transcribed, but not translated. UTR sequences diverge more rapidly during evolution than structural regions.
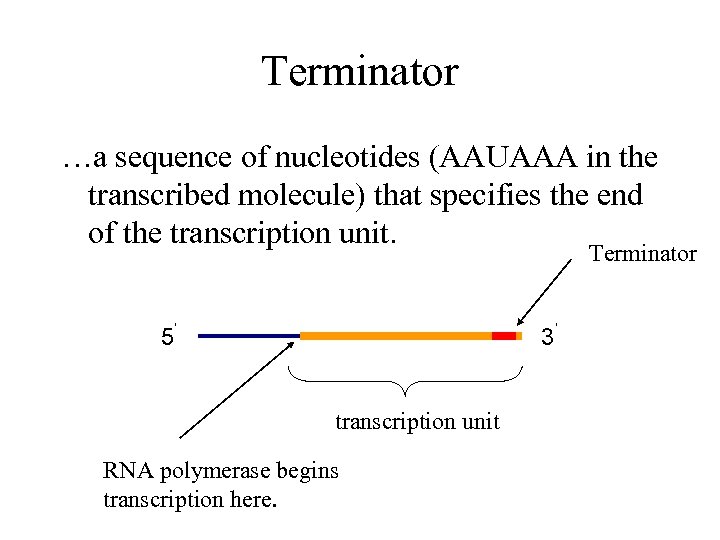 Terminator …a sequence of nucleotides (AAUAAA in the transcribed molecule) that specifies the end of the transcription unit. Terminator 5’ 3’ transcription unit RNA polymerase begins transcription here.
Terminator …a sequence of nucleotides (AAUAAA in the transcribed molecule) that specifies the end of the transcription unit. Terminator 5’ 3’ transcription unit RNA polymerase begins transcription here.
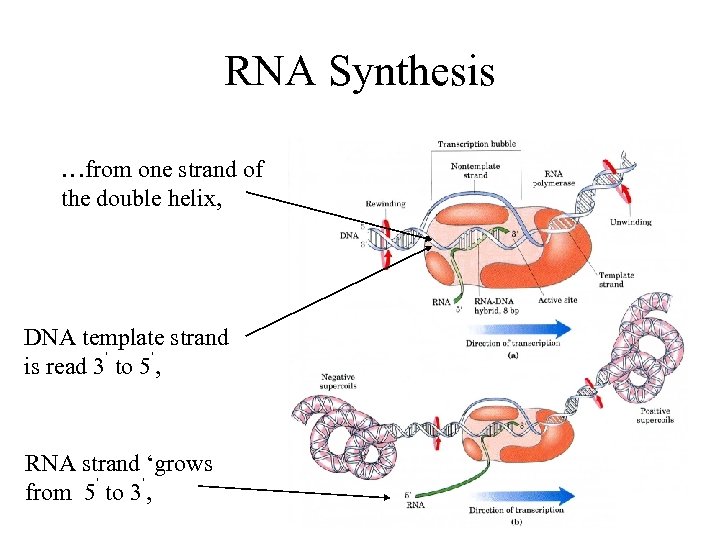 RNA Synthesis …from one strand of the double helix, DNA template strand is read 3’ to 5’, RNA strand ‘grows from 5’ to 3’,
RNA Synthesis …from one strand of the double helix, DNA template strand is read 3’ to 5’, RNA strand ‘grows from 5’ to 3’,
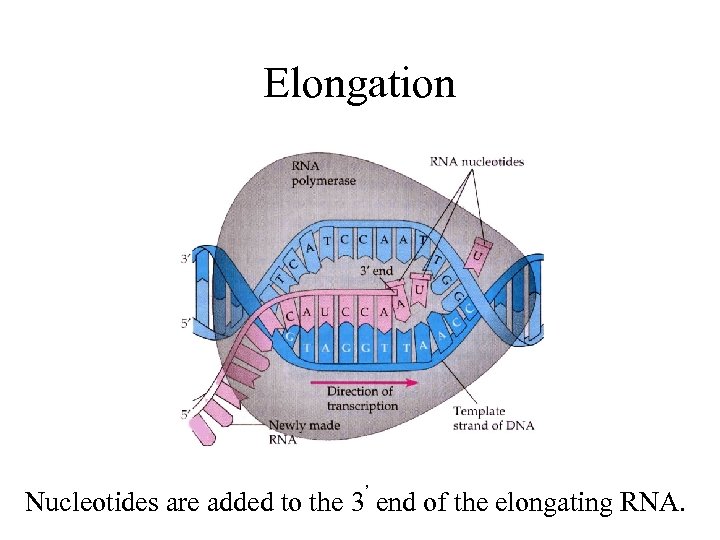 Elongation Nucleotides are added to the 3’ end of the elongating RNA.
Elongation Nucleotides are added to the 3’ end of the elongating RNA.
 m. RNA vs. pre-m. RNA • prokaryotic m. RNA synthesis described so far requires little, or no further modification prior to translation into proteins, • eukaryotic m. RNA requires extensive modifications.
m. RNA vs. pre-m. RNA • prokaryotic m. RNA synthesis described so far requires little, or no further modification prior to translation into proteins, • eukaryotic m. RNA requires extensive modifications.
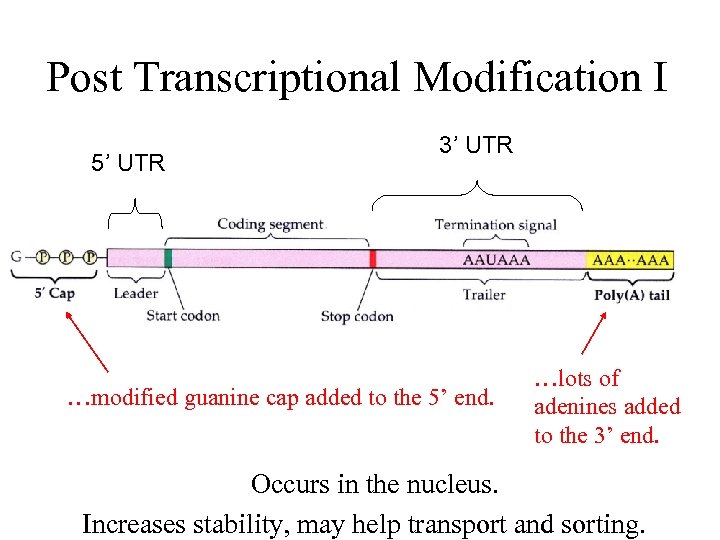 Post Transcriptional Modification I 5’ UTR 3’ UTR …modified guanine cap added to the 5’ end. …lots of adenines added to the 3’ end. Occurs in the nucleus. Increases stability, may help transport and sorting.
Post Transcriptional Modification I 5’ UTR 3’ UTR …modified guanine cap added to the 5’ end. …lots of adenines added to the 3’ end. Occurs in the nucleus. Increases stability, may help transport and sorting.
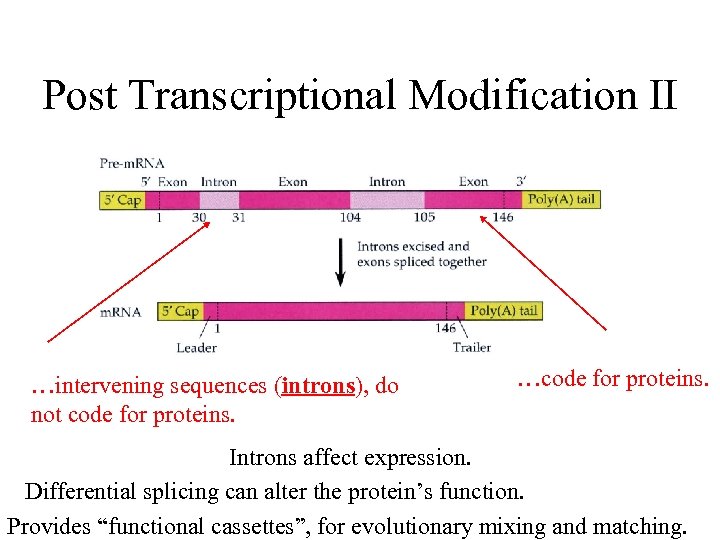 Post Transcriptional Modification II …intervening sequences (introns), do not code for proteins. …code for proteins. Introns affect expression. Differential splicing can alter the protein’s function. Provides “functional cassettes”, for evolutionary mixing and matching.
Post Transcriptional Modification II …intervening sequences (introns), do not code for proteins. …code for proteins. Introns affect expression. Differential splicing can alter the protein’s function. Provides “functional cassettes”, for evolutionary mixing and matching.
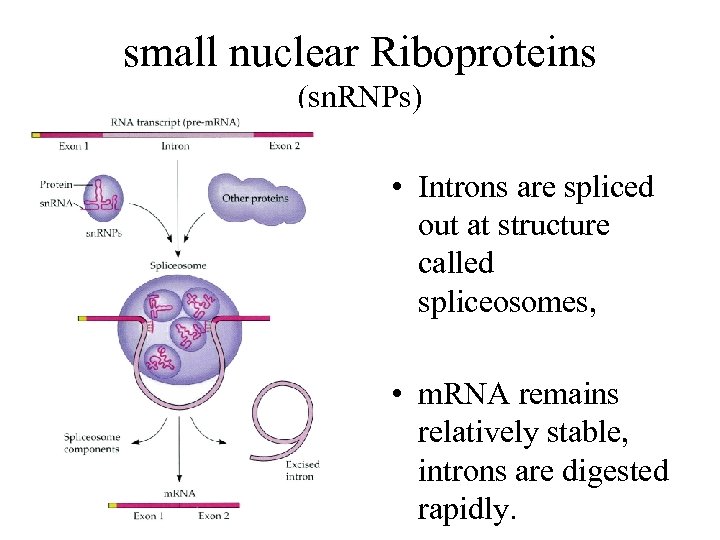 small nuclear Riboproteins (sn. RNPs) • Introns are spliced out at structure called spliceosomes, • m. RNA remains relatively stable, introns are digested rapidly.
small nuclear Riboproteins (sn. RNPs) • Introns are spliced out at structure called spliceosomes, • m. RNA remains relatively stable, introns are digested rapidly.
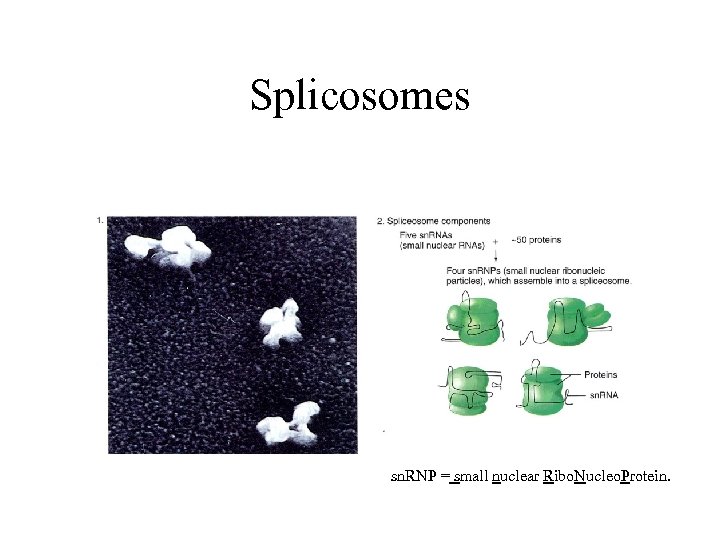 Splicosomes sn. RNP = small nuclear Ribo. Nucleo. Protein.
Splicosomes sn. RNP = small nuclear Ribo. Nucleo. Protein.
 Study Figure 9. 15 for Wednesday
Study Figure 9. 15 for Wednesday
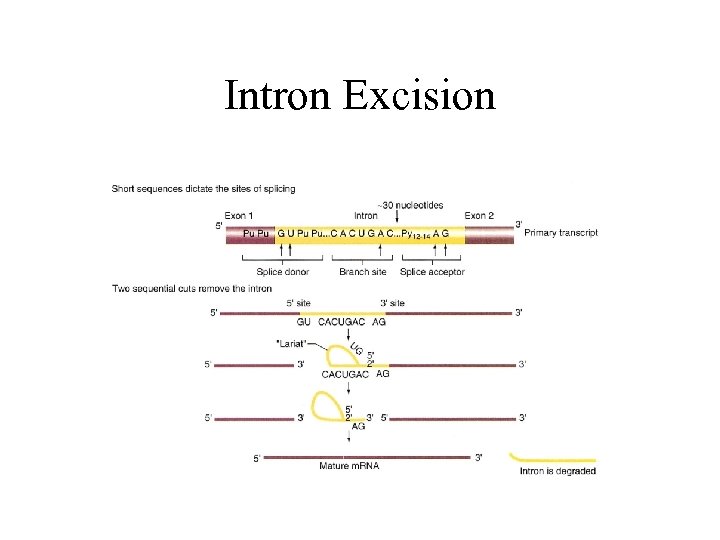 Intron Excision
Intron Excision
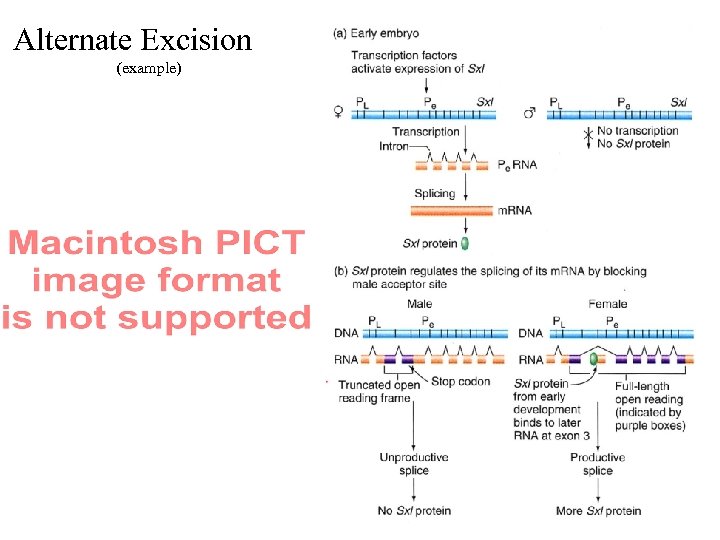 Alternate Excision (example)
Alternate Excision (example)
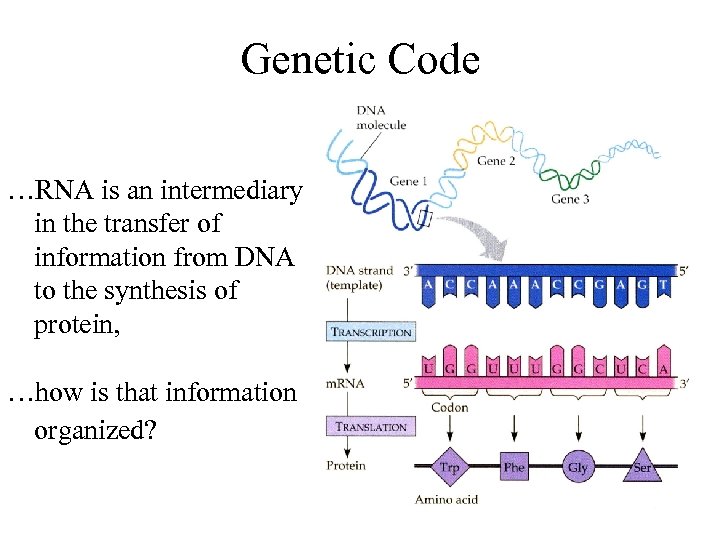 Genetic Code …RNA is an intermediary in the transfer of information from DNA to the synthesis of protein, …how is that information organized?
Genetic Code …RNA is an intermediary in the transfer of information from DNA to the synthesis of protein, …how is that information organized?
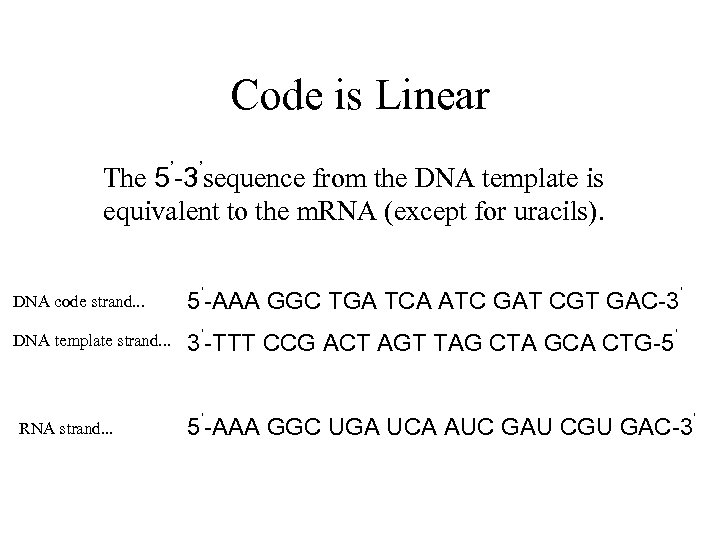 Code is Linear The 5’-3’sequence from the DNA template is equivalent to the m. RNA (except for uracils). DNA code strand. . . 5’-AAA GGC TGA TCA ATC GAT CGT GAC-3’ DNA template strand. . . 3’-TTT CCG ACT AGT TAG CTA GCA CTG-5’ RNA strand. . . 5’-AAA GGC UGA UCA AUC GAU CGU GAC-3’
Code is Linear The 5’-3’sequence from the DNA template is equivalent to the m. RNA (except for uracils). DNA code strand. . . 5’-AAA GGC TGA TCA ATC GAT CGT GAC-3’ DNA template strand. . . 3’-TTT CCG ACT AGT TAG CTA GCA CTG-5’ RNA strand. . . 5’-AAA GGC UGA UCA AUC GAU CGU GAC-3’
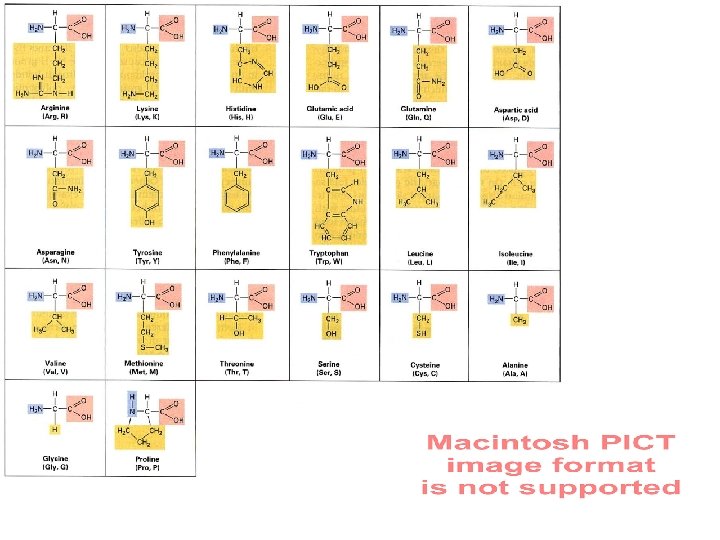
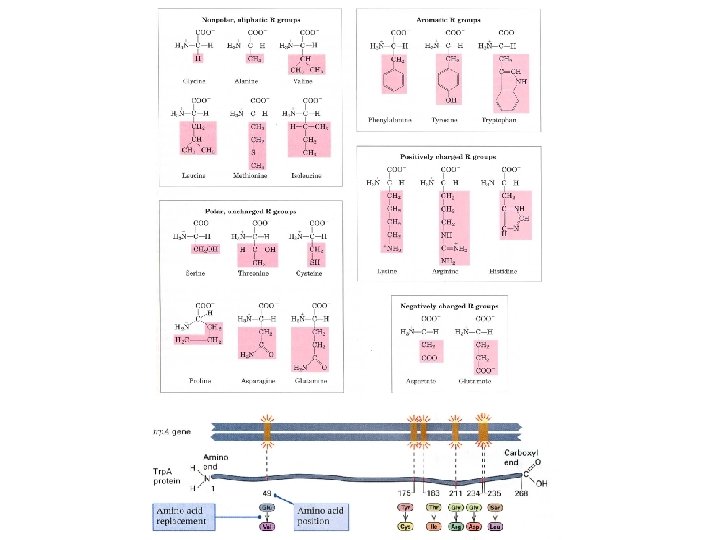
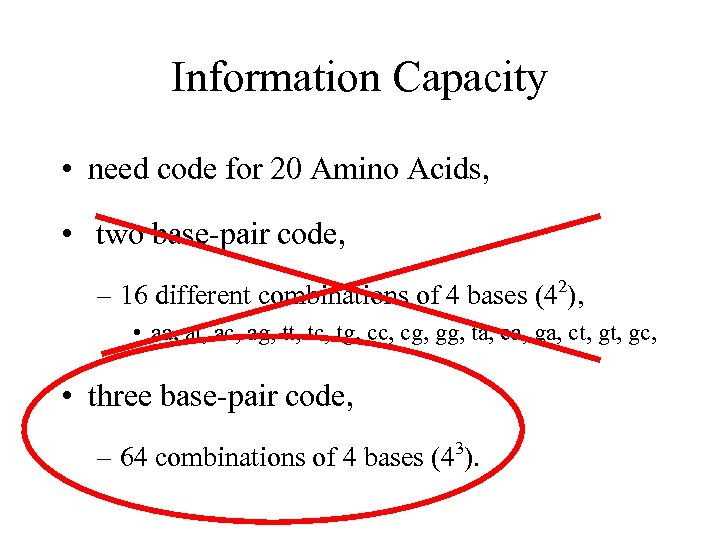 Information Capacity • need code for 20 Amino Acids, • two base-pair code, 2 – 16 different combinations of 4 bases (4 ), • aa, at, ac, ag, tt, tc, tg, cc, cg, gg, ta, ca, ga, ct, gc, • three base-pair code, – 64 combinations of 4 bases (43).
Information Capacity • need code for 20 Amino Acids, • two base-pair code, 2 – 16 different combinations of 4 bases (4 ), • aa, at, ac, ag, tt, tc, tg, cc, cg, gg, ta, ca, ga, ct, gc, • three base-pair code, – 64 combinations of 4 bases (43).
 Codons …a triplet of nucleotide bases that specifies or encodes the information for a specific amino acid, – also need codons to indicate the beginning and end of the protein to be synthesized.
Codons …a triplet of nucleotide bases that specifies or encodes the information for a specific amino acid, – also need codons to indicate the beginning and end of the protein to be synthesized.
 r. II Again Revertants returned the code to an “in frame” conditions.
r. II Again Revertants returned the code to an “in frame” conditions.
 Code is Degenerate 20 amino acid codons + start and stop codons 20 -some required 64 possible All combinations are used.
Code is Degenerate 20 amino acid codons + start and stop codons 20 -some required 64 possible All combinations are used.
 Start/Stop Codons …AUG codes for the ‘start of translation’, a methionine, – most proteins thus begin with the amino acid methionine, …UAA, UAG and UGA are stop codons, indicating the C terminus of the protein.
Start/Stop Codons …AUG codes for the ‘start of translation’, a methionine, – most proteins thus begin with the amino acid methionine, …UAA, UAG and UGA are stop codons, indicating the C terminus of the protein.
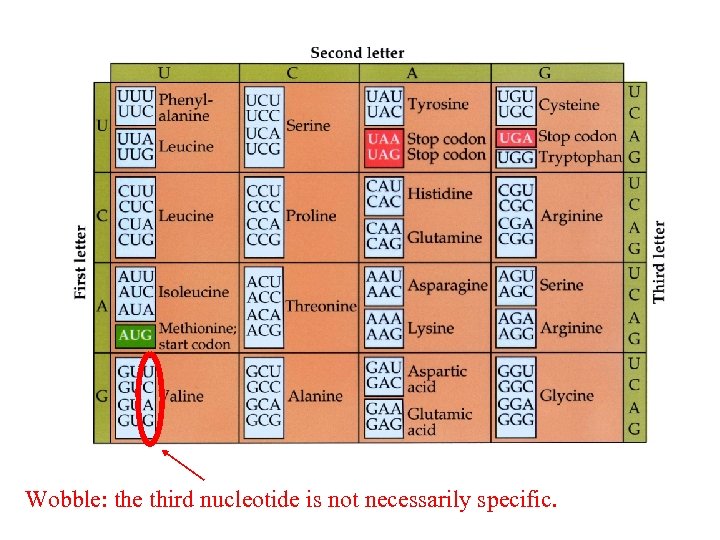 Wobble: the third nucleotide is not necessarily specific.
Wobble: the third nucleotide is not necessarily specific.
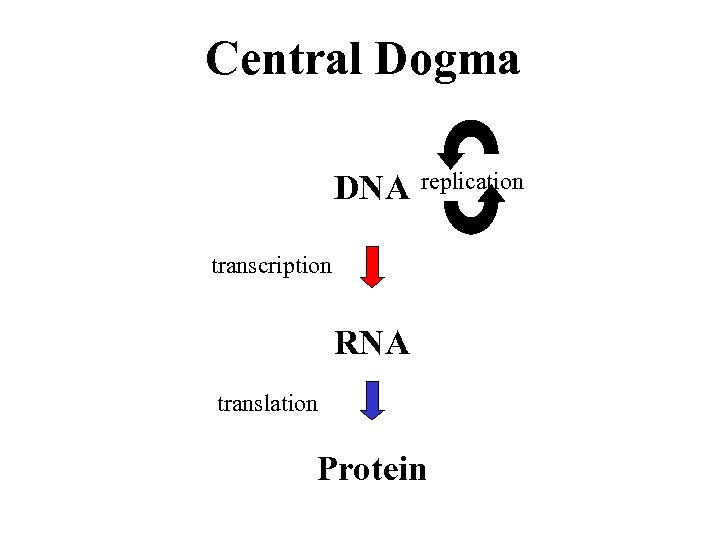 Central Dogma DNA replication transcription RNA translation Protein
Central Dogma DNA replication transcription RNA translation Protein
 Translation …the synthesis of a polypeptide. This occurs on ribosomes using the information encoded on m. RNA, – t. RNA molecules mediate the transfer of information between m. RNA and the growing polypeptide.
Translation …the synthesis of a polypeptide. This occurs on ribosomes using the information encoded on m. RNA, – t. RNA molecules mediate the transfer of information between m. RNA and the growing polypeptide.
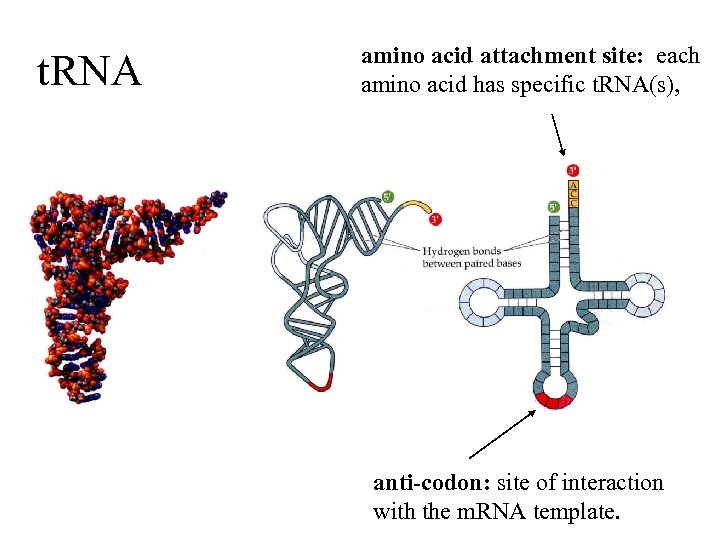 t. RNA amino acid attachment site: each amino acid has specific t. RNA(s), anti-codon: site of interaction with the m. RNA template.
t. RNA amino acid attachment site: each amino acid has specific t. RNA(s), anti-codon: site of interaction with the m. RNA template.
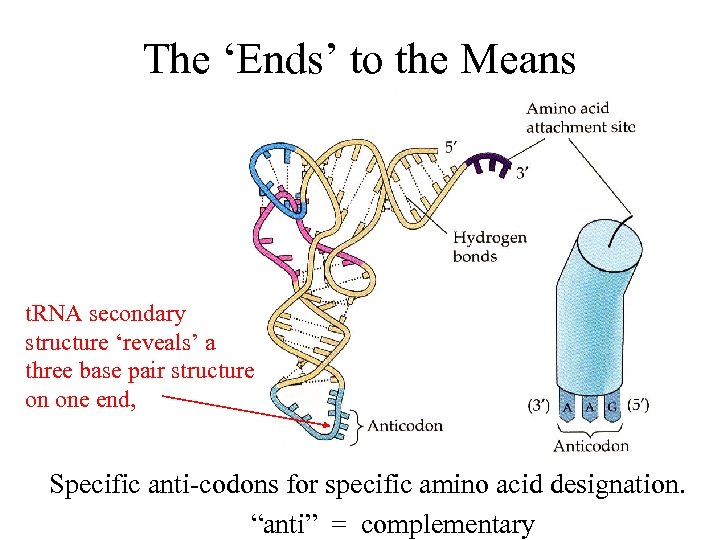 The ‘Ends’ to the Means t. RNA secondary structure ‘reveals’ a three base pair structure on one end, Specific anti-codons for specific amino acid designation. “anti” = complementary
The ‘Ends’ to the Means t. RNA secondary structure ‘reveals’ a three base pair structure on one end, Specific anti-codons for specific amino acid designation. “anti” = complementary
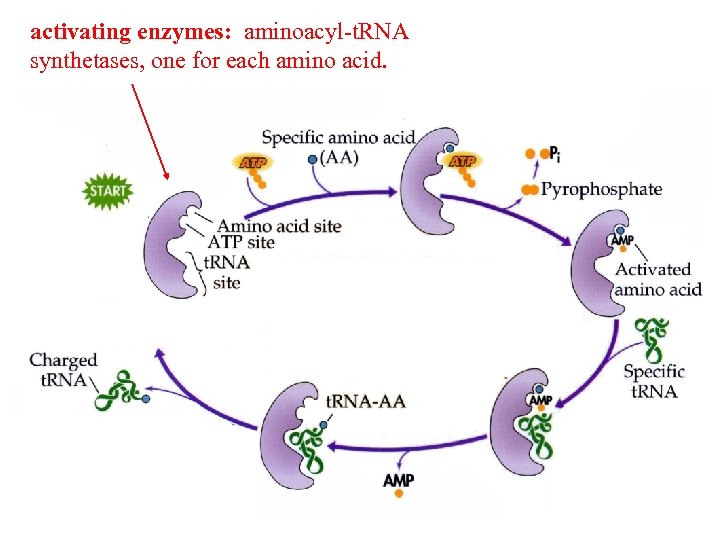 activating enzymes: aminoacyl-t. RNA synthetases, one for each amino acid.
activating enzymes: aminoacyl-t. RNA synthetases, one for each amino acid.
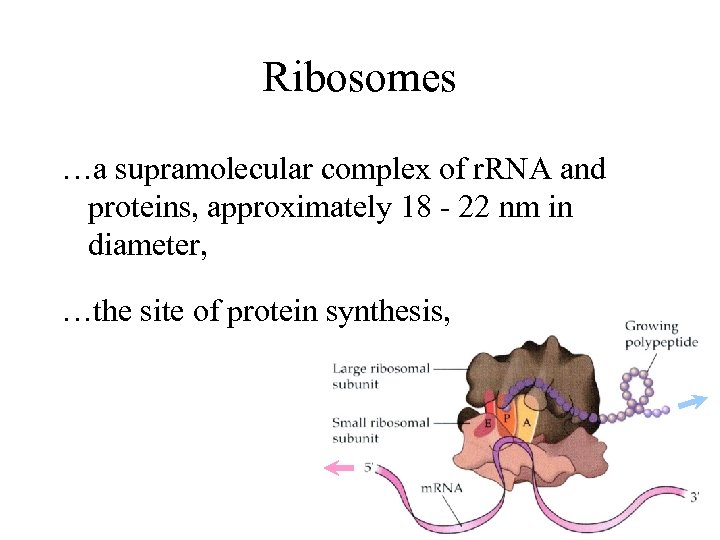 Ribosomes …a supramolecular complex of r. RNA and proteins, approximately 18 - 22 nm in diameter, …the site of protein synthesis,
Ribosomes …a supramolecular complex of r. RNA and proteins, approximately 18 - 22 nm in diameter, …the site of protein synthesis,
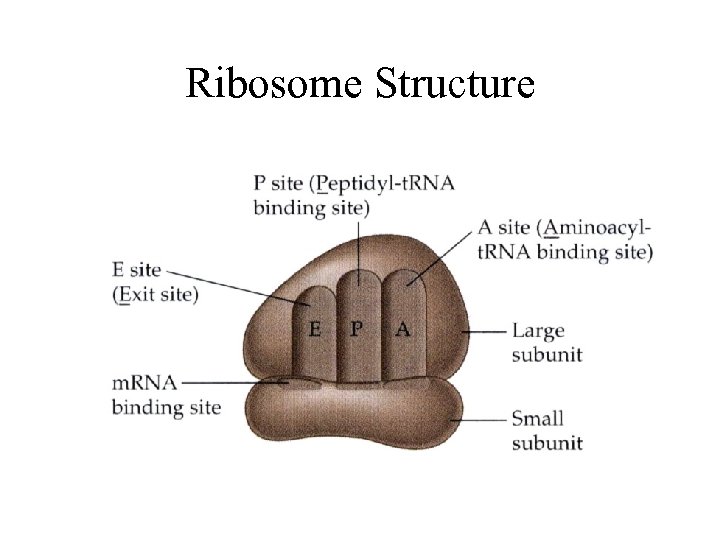 Ribosome Structure
Ribosome Structure
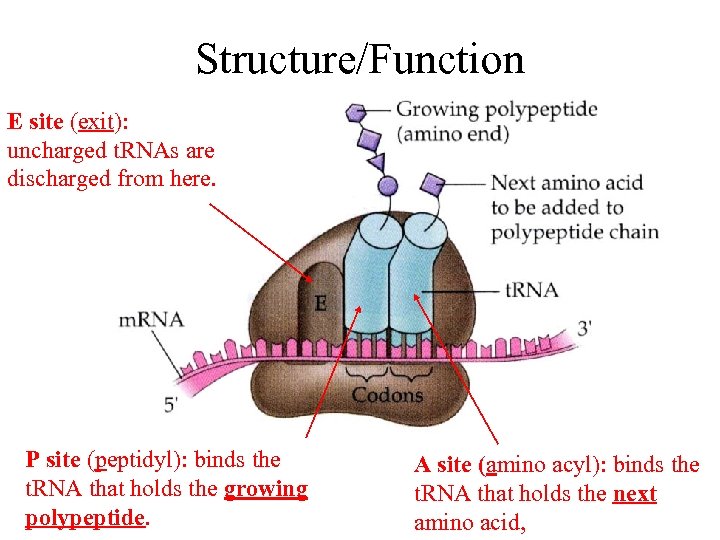 Structure/Function E site (exit): uncharged t. RNAs are discharged from here. P site (peptidyl): binds the t. RNA that holds the growing polypeptide. A site (amino acyl): binds the t. RNA that holds the next amino acid,
Structure/Function E site (exit): uncharged t. RNAs are discharged from here. P site (peptidyl): binds the t. RNA that holds the growing polypeptide. A site (amino acyl): binds the t. RNA that holds the next amino acid,
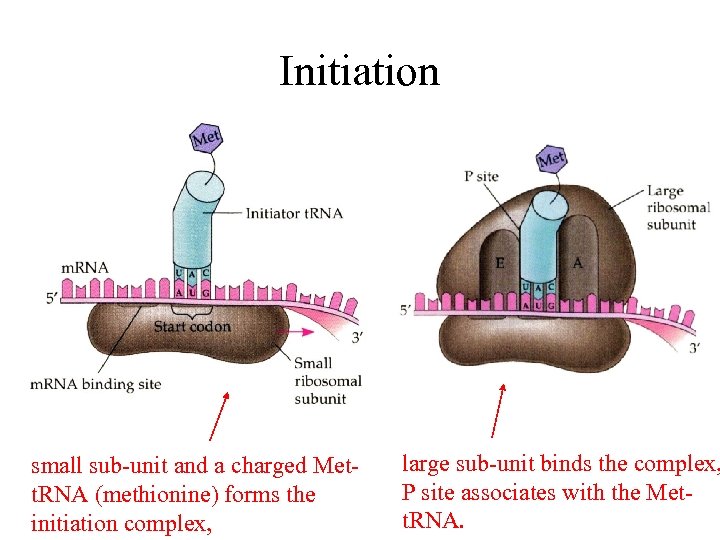 Initiation small sub-unit and a charged Mett. RNA (methionine) forms the initiation complex, large sub-unit binds the complex, P site associates with the Mett. RNA.
Initiation small sub-unit and a charged Mett. RNA (methionine) forms the initiation complex, large sub-unit binds the complex, P site associates with the Mett. RNA.
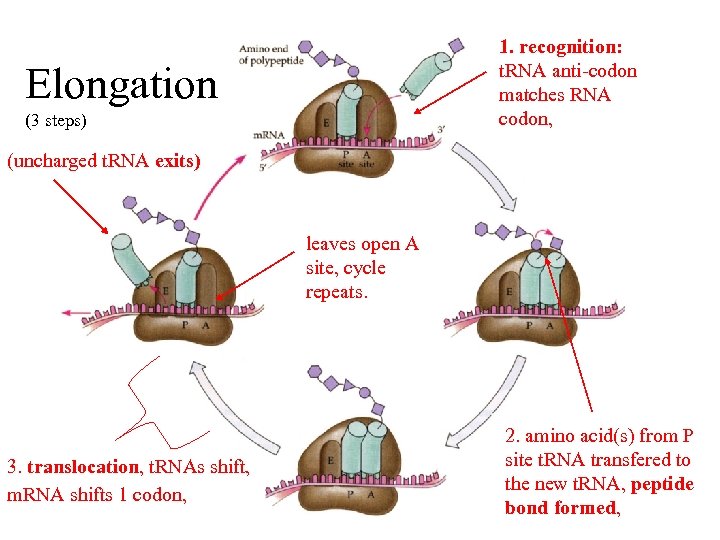 1. recognition: t. RNA anti-codon matches RNA codon, Elongation (3 steps) (uncharged t. RNA exits) leaves open A site, cycle repeats. 3. translocation, t. RNAs shift, m. RNA shifts 1 codon, 2. amino acid(s) from P site t. RNA transfered to the new t. RNA, peptide bond formed,
1. recognition: t. RNA anti-codon matches RNA codon, Elongation (3 steps) (uncharged t. RNA exits) leaves open A site, cycle repeats. 3. translocation, t. RNAs shift, m. RNA shifts 1 codon, 2. amino acid(s) from P site t. RNA transfered to the new t. RNA, peptide bond formed,
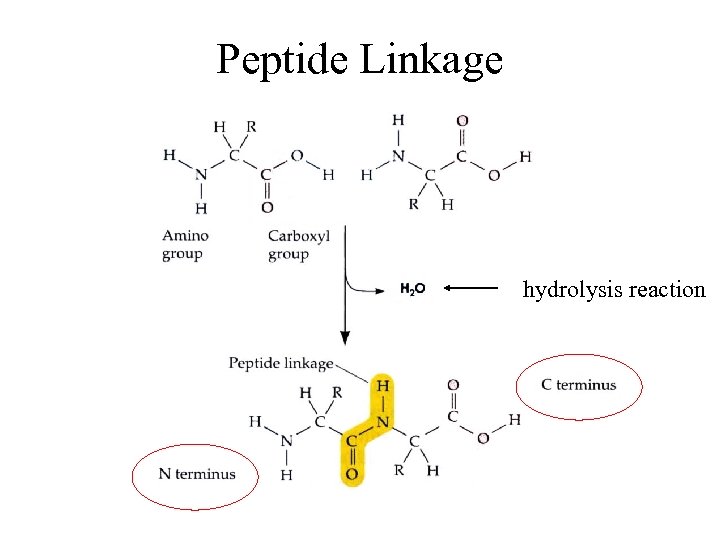 Peptide Linkage hydrolysis reaction
Peptide Linkage hydrolysis reaction
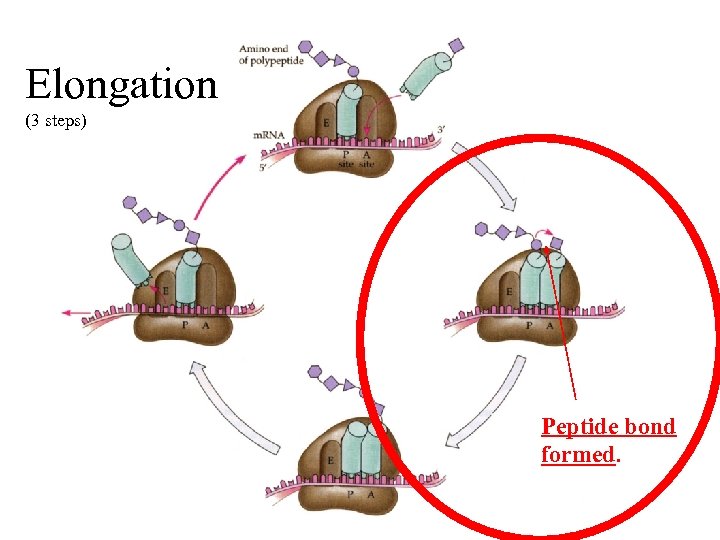 Elongation (3 steps) Peptide bond formed.
Elongation (3 steps) Peptide bond formed.
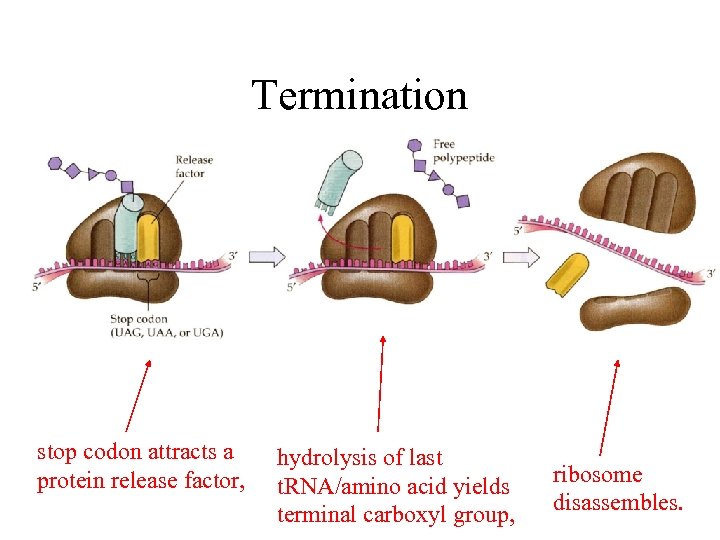 Termination stop codon attracts a protein release factor, hydrolysis of last t. RNA/amino acid yields terminal carboxyl group, ribosome disassembles.
Termination stop codon attracts a protein release factor, hydrolysis of last t. RNA/amino acid yields terminal carboxyl group, ribosome disassembles.
 N-Terminus --> C-Terminus. . . polypeptides are synthesized beginning from the N-terminus (amino terminus) and going to the C-terminus (carboxy terminus), ’ ’ …this corresponds to the 5 -3 DNA Coding sequence.
N-Terminus --> C-Terminus. . . polypeptides are synthesized beginning from the N-terminus (amino terminus) and going to the C-terminus (carboxy terminus), ’ ’ …this corresponds to the 5 -3 DNA Coding sequence.
 Wednesday
Wednesday


