d199feb5c77224ea2a63d413de5998e2.ppt
- Количество слайдов: 51

The Uses of Object Shape from Images in Medicine Stephen M. Pizer Kenan Professor Medical Image Display & Analysis Group University of North Carolina Credits: Many on MIDAG, especially Daniel Fritsch, Guido Gerig, Edward Chaney, Elizabeth Bullitt, Stephen Aylward, George Stetten, Gregg Tracton, Tom Fletcher, Andrew Thall, Paul Yushkevich, Nikki Levine, Greg Clary, David Chen MIDAG@UNC
![Object Representation in Medical Image Analysis ä Extract an object from image(s) [segmentation] ä Object Representation in Medical Image Analysis ä Extract an object from image(s) [segmentation] ä](https://present5.com/presentation/d199feb5c77224ea2a63d413de5998e2/image-2.jpg)
Object Representation in Medical Image Analysis ä Extract an object from image(s) [segmentation] ä Radiotherapy Tumor; plan to hit it ä Radiosensitive normal anatomy; plan to miss it ä PD MRA T 2 T 1 Contrast ä Surgery Plan to remove it ä Plan to miss it ä During surgery, view where it is & effect of treatment ä ä Radiology ä View it to judge its pathology MIDAG@UNC
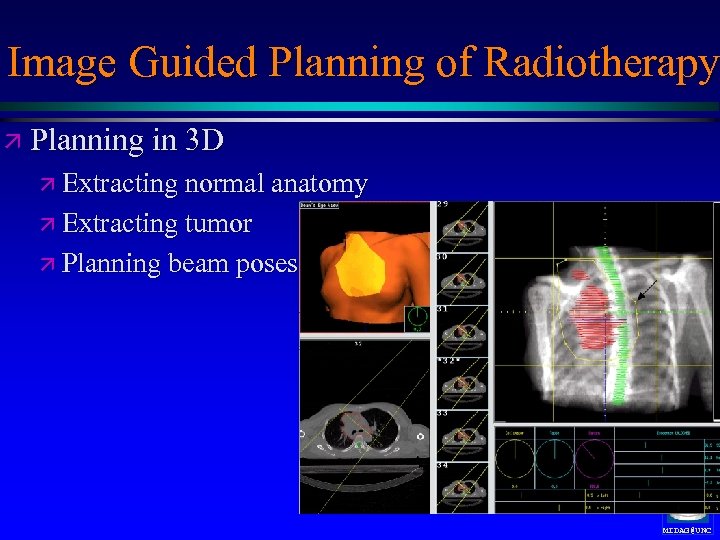
Image Guided Planning of Radiotherapy ä Planning in 3 D ä Extracting normal anatomy ä Extracting tumor ä Planning beam poses MIDAG@UNC
Object Representation in Medical Image Analysis ä Registration (find geometric transformation that brings two images into alignment) ä Radiotherapy ä Fuse multimodality images (3 D/3 D) for planning ä Verify patient placement (3 D/2 D) ä Surgery ä Fuse multimodality images (3 D/3 D or 2 D) for planning ä Fuse preoperative (3 D) & intraoperative (2 D) images ä Radiology ä Fuse multimodality images (3 D/3 D) for diagnosis MIDAG@UNC
Object Representation in Medical Image Analysis ä Shape & Volume Measurement ä Make physical measurement ä Radiotherapy ä Measure effect of therapy on tumor ä Radiology, ä Neurosciences Use measurement in science of object development ä Find how probable an object is ä Radiology, Neurosciences Use measurement as quantitative input to diagnosis ä Use measurement in science of object development ä ä Use as prior in object extraction ä E. g. , extract the kidney shaped object MIDAG@UNC
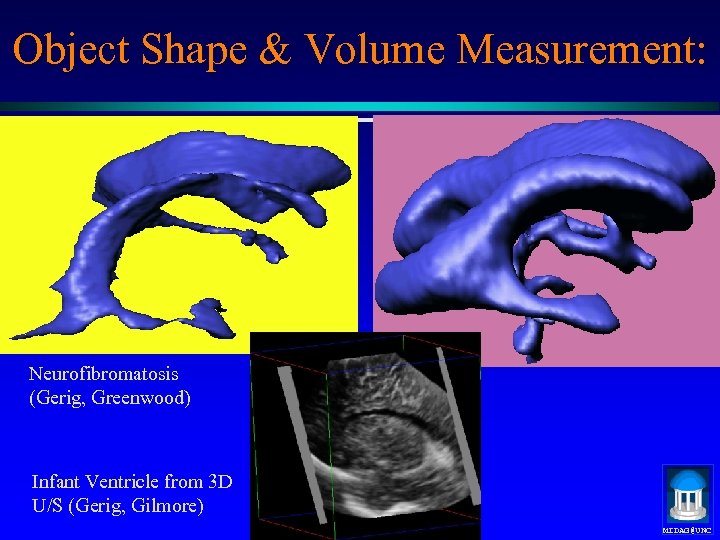
Object Shape & Volume Measurement: Neurofibromatosis (Gerig, Greenwood) Infant Ventricle from 3 D U/S (Gerig, Gilmore) MIDAG@UNC
Object Extraction (Segmentation) ä Approach 1: preanalyze, then fit to model ä Neurosurgery (MR Angiogram), Radiology (CT) ä Vessels, ribs, bronchi, bowel via tube skeletons ä Cardiology (3 D Ultrasound) Geometry via clouds of medial atoms ä Fit appropriately labeled clouds to 3 D LV model ä ä Cardiac Nuclear Medicine (2 D Gated Blood Pool Cine) Extract LV, with previous frame providing model ä Extraction via deformable m-rep model ä Shape from extracted LV; analyze shape series ä ä Surgery, Radiation Oncology (Multimodality MRI) ä Extract tumor, using local shape characteristics MIDAG@UNC
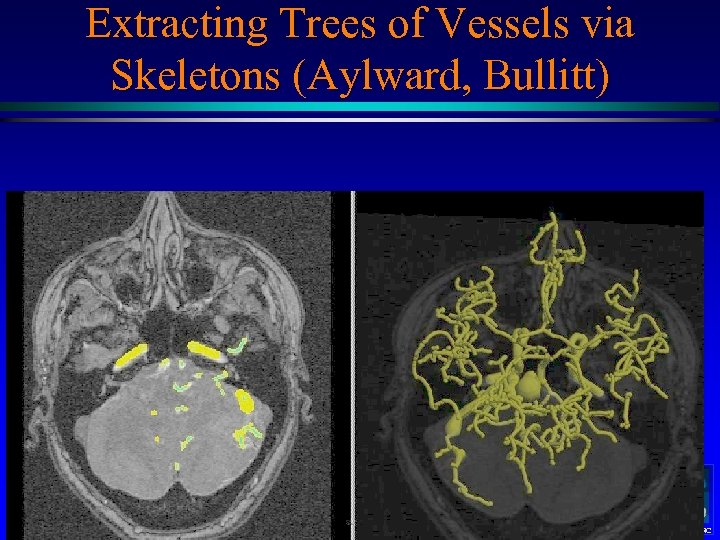
Extracting Trees of Vessels via Skeletons (Aylward, Bullitt) MIDAG@UNC
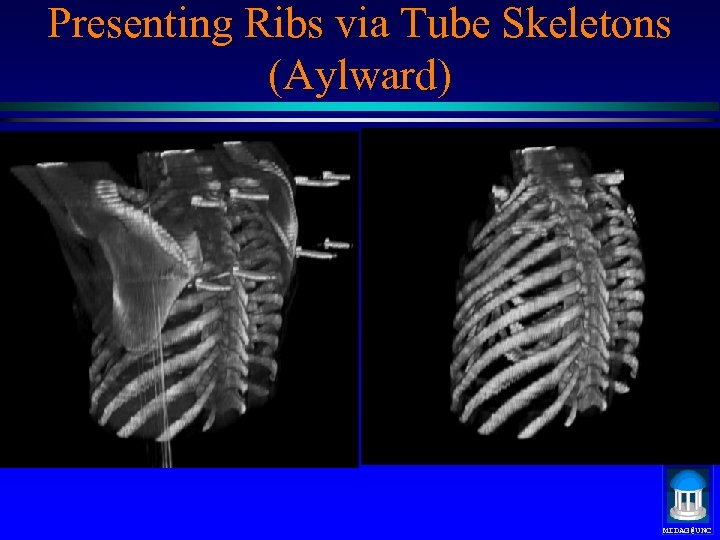
Presenting Ribs via Tube Skeletons (Aylward) MIDAG@UNC
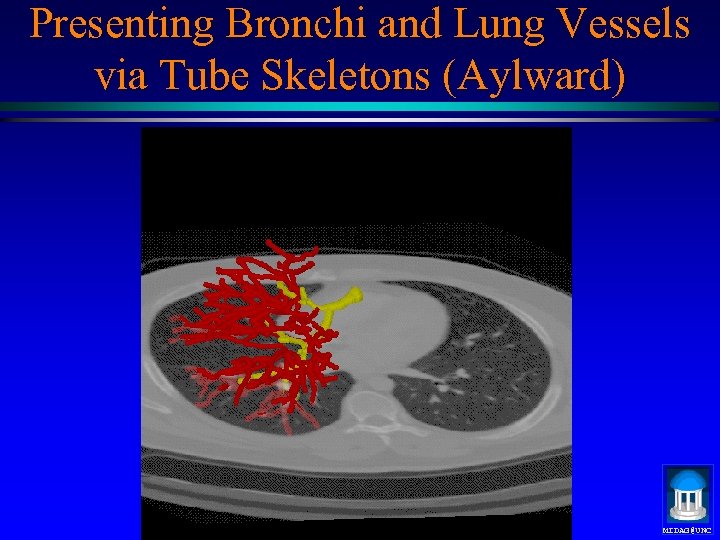
Presenting Bronchi and Lung Vessels via Tube Skeletons (Aylward) MIDAG@UNC
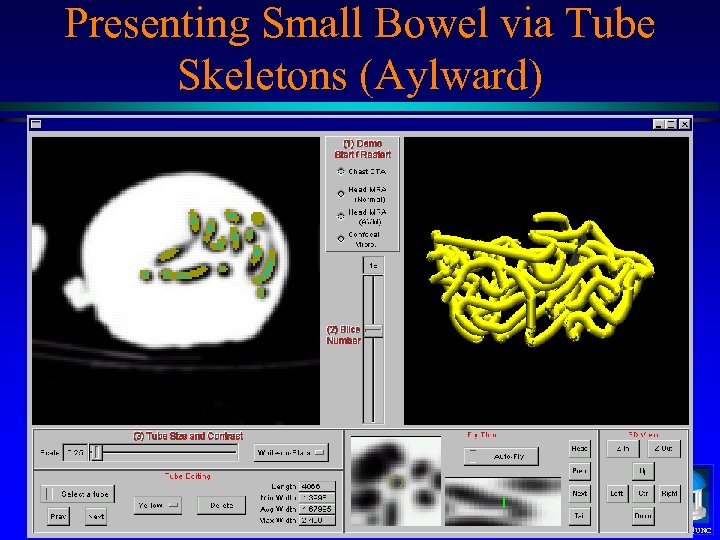
Presenting Small Bowel via Tube Skeletons (Aylward) MIDAG@UNC
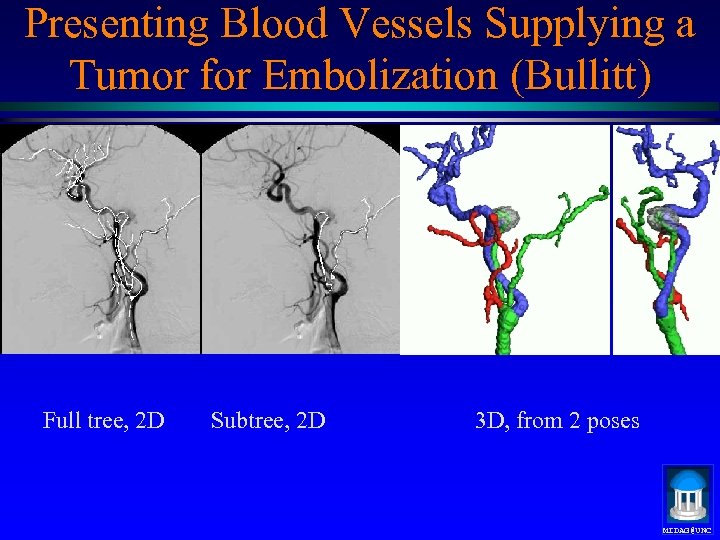
Presenting Blood Vessels Supplying a Tumor for Embolization (Bullitt) Full tree, 2 D Subtree, 2 D 3 D, from 2 poses MIDAG@UNC
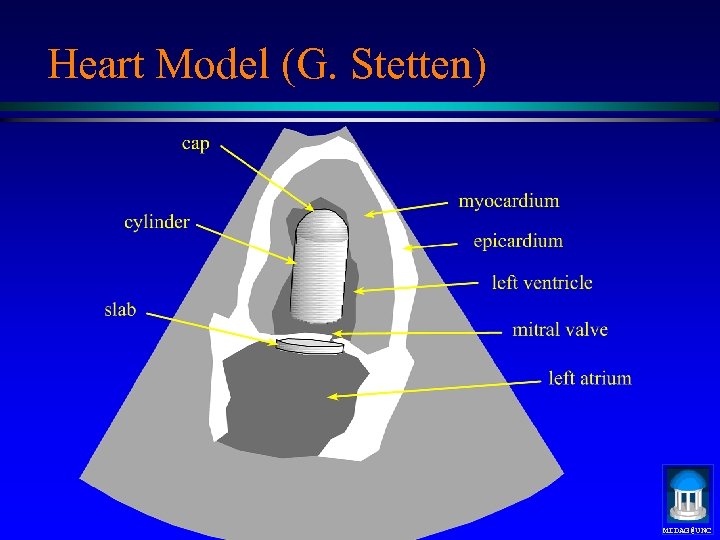
Heart Model (G. Stetten) MIDAG@UNC
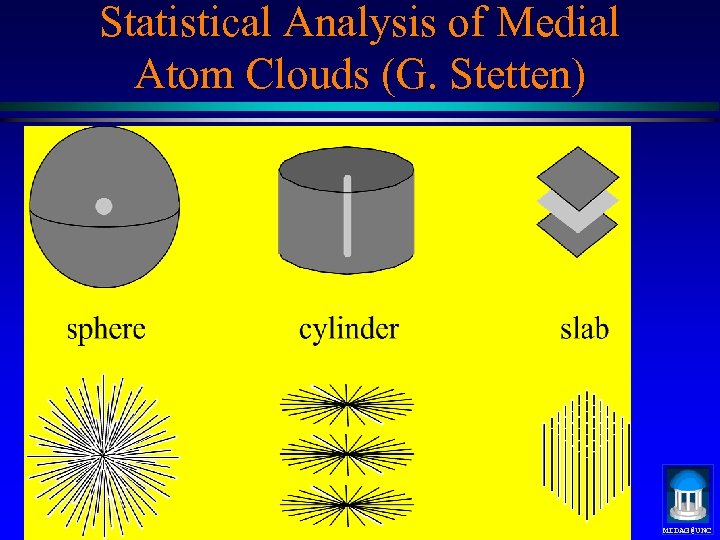
Statistical Analysis of Medial Atom Clouds (G. Stetten) MIDAG@UNC
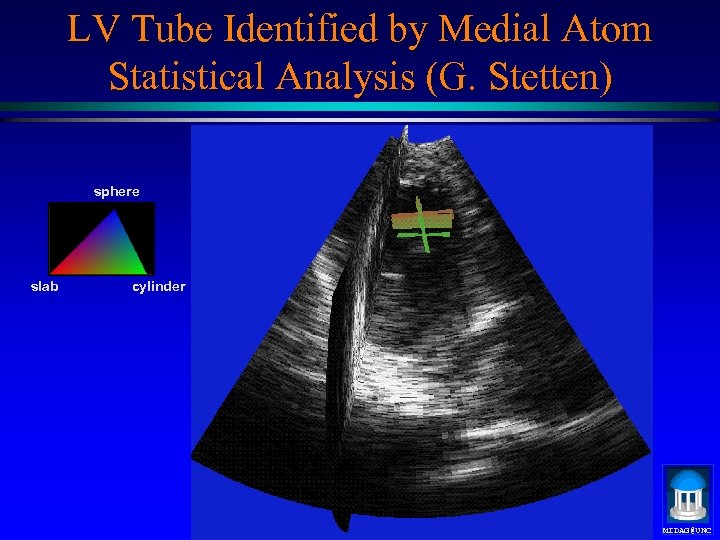
LV Tube Identified by Medial Atom Statistical Analysis (G. Stetten) sphere slab cylinder MIDAG@UNC
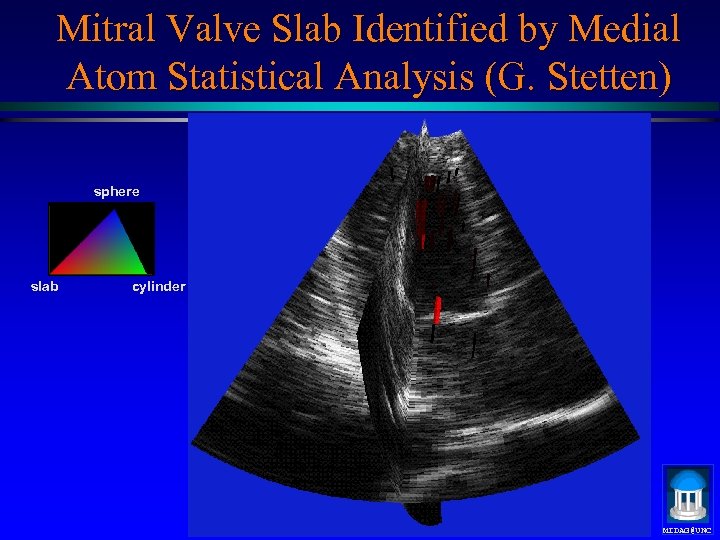
Mitral Valve Slab Identified by Medial Atom Statistical Analysis (G. Stetten) sphere slab cylinder MIDAG@UNC
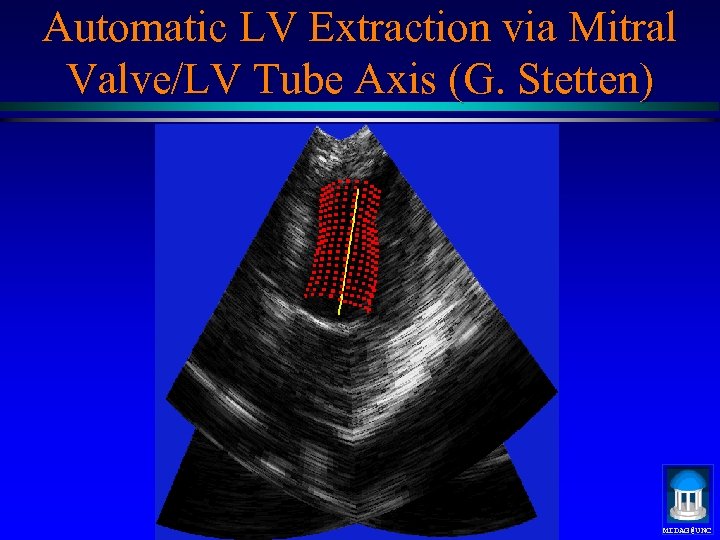
Automatic LV Extraction via Mitral Valve/LV Tube Axis (G. Stetten) MIDAG@UNC
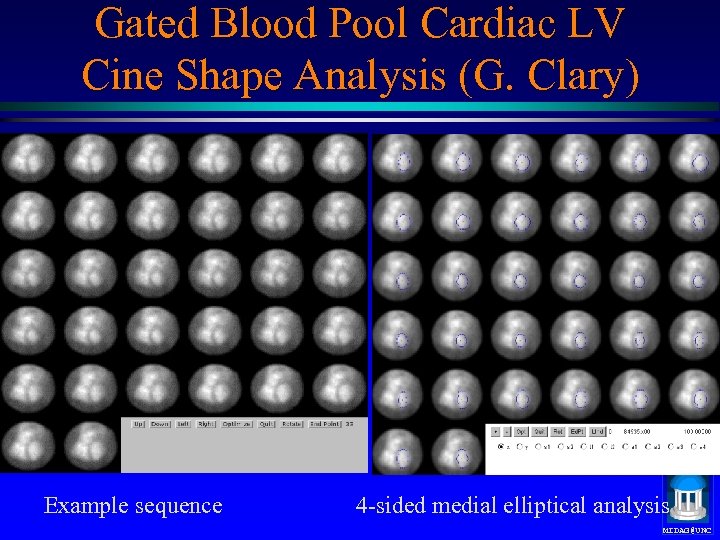
Gated Blood Pool Cardiac LV Cine Shape Analysis (G. Clary) Example sequence 4 -sided medial elliptical analysis MIDAG@UNC
Object Extraction (Segmentation) ä Approach 2: deform model to optimize reward for image match + reward for shape normality ä Radiation Oncology (CT or MRI) ä Abdominal, pelvic organs ä Deform m-reps model ä Neurosciences (MRI or 3 D Ultrasound) ä Internal brain structures ä Spherical harmonics boundary model ä Deformable m-reps model ä Neurosurgery (CT) ä Vertebrae MIDAG@UNC
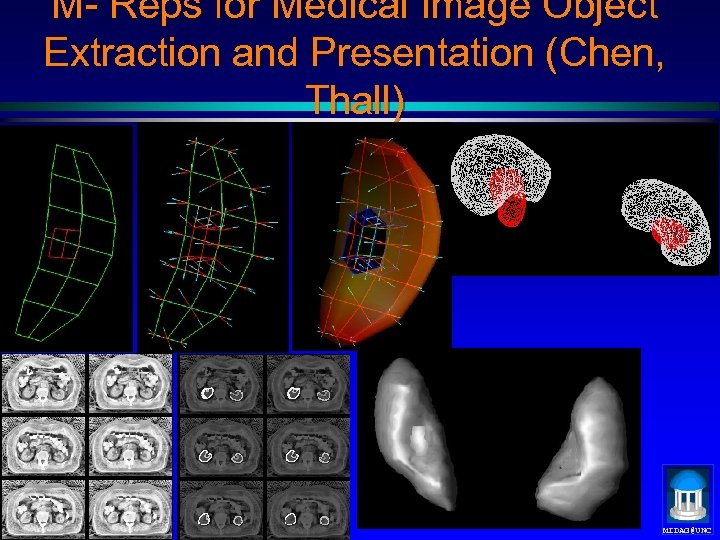
M- Reps for Medical Image Object Extraction and Presentation (Chen, Thall) MIDAG@UNC
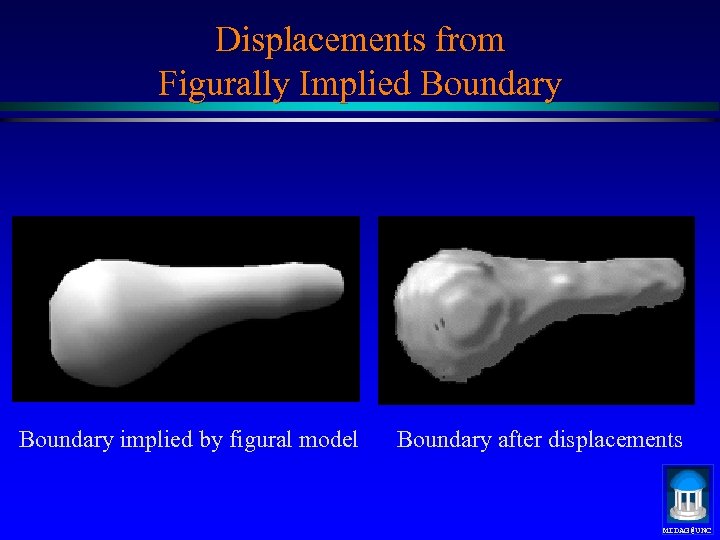
Displacements from Figurally Implied Boundary implied by figural model Boundary after displacements MIDAG@UNC
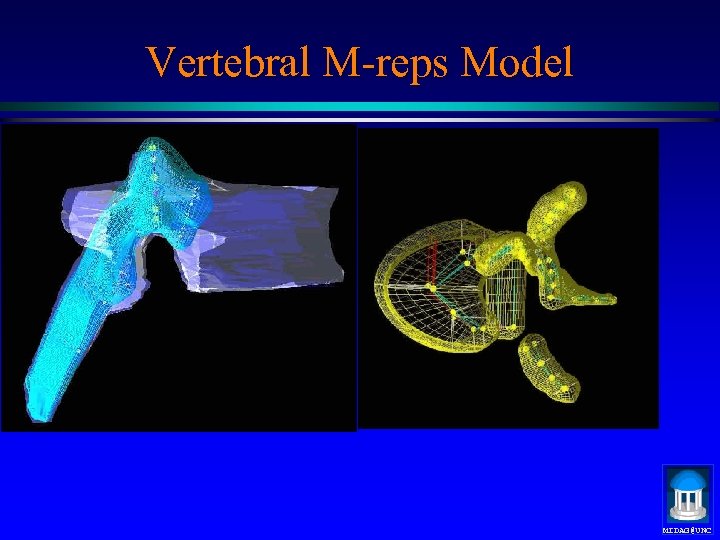
Vertebral M-reps Model MIDAG@UNC
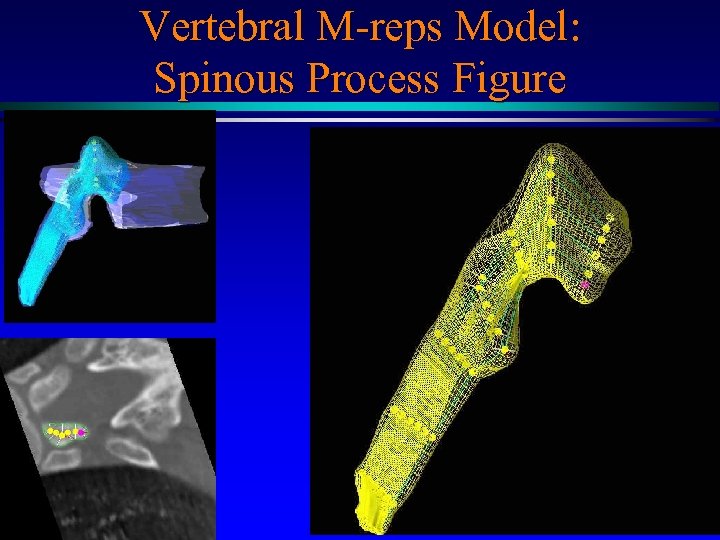
Vertebral M-reps Model: Spinous Process Figure MIDAG@UNC
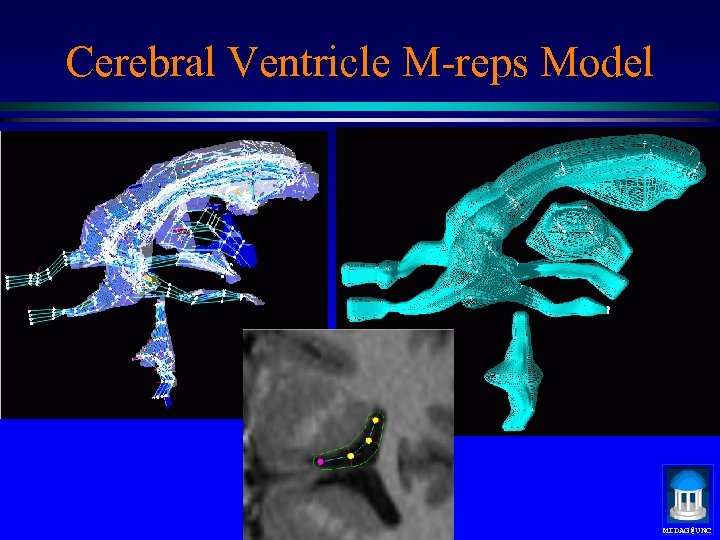
Cerebral Ventricle M-reps Model MIDAG@UNC
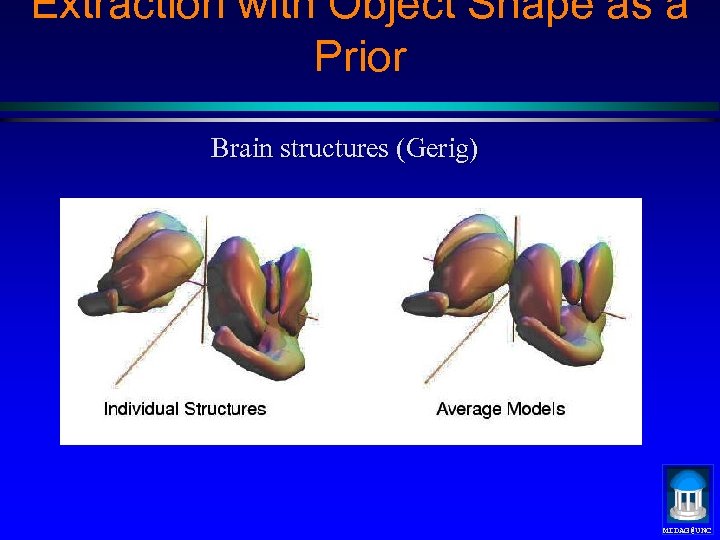
Extraction with Object Shape as a Prior Brain structures (Gerig) MIDAG@UNC
Registration ä Registration (find geometric transformation that brings two images into alignment) ä Radiotherapy ä Fuse multimodality images (3 D/3 D) for planning ä Verify patient placement (3 D/2 D) ä Surgery ä Fuse multimodality images (3 D/3 D or 2 D) for planning ä Fuse preoperative (3 D) & intraoperative (2 D) images ä Radiology ä Fuse multimodality images (3 D/3 D) for diagnosis MIDAG@UNC
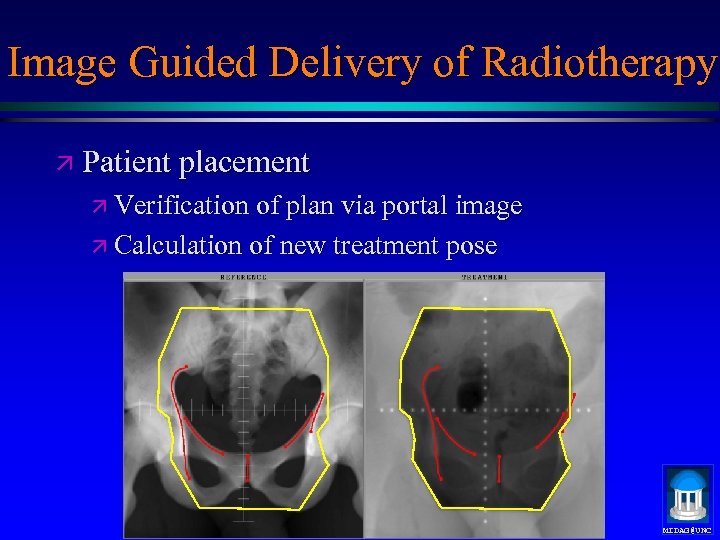
Image Guided Delivery of Radiotherapy ä Patient placement ä Verification of plan via portal image ä Calculation of new treatment pose MIDAG@UNC
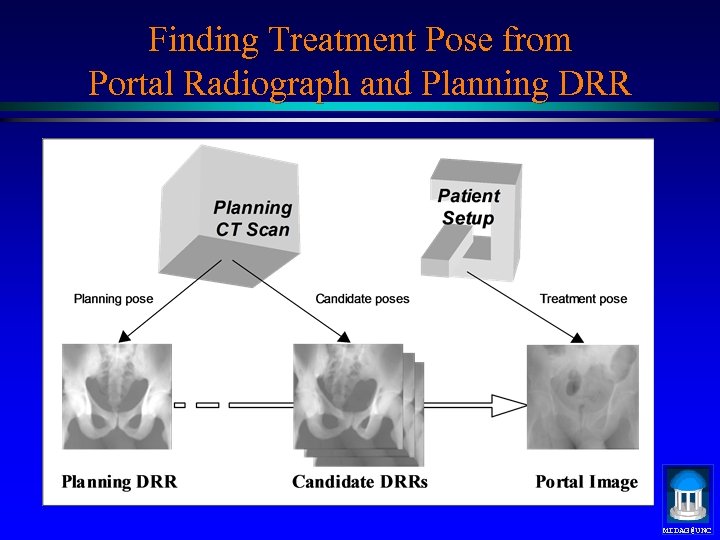
Finding Treatment Pose from Portal Radiograph and Planning DRR MIDAG@UNC
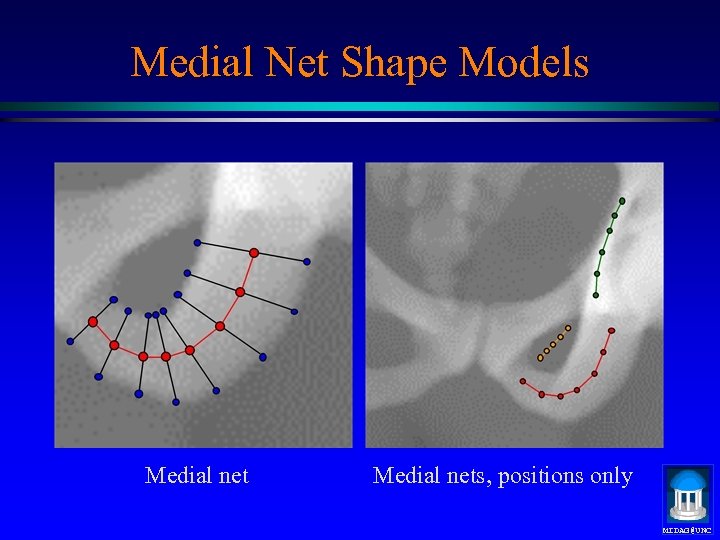
Medial Net Shape Models Medial nets, positions only MIDAG@UNC
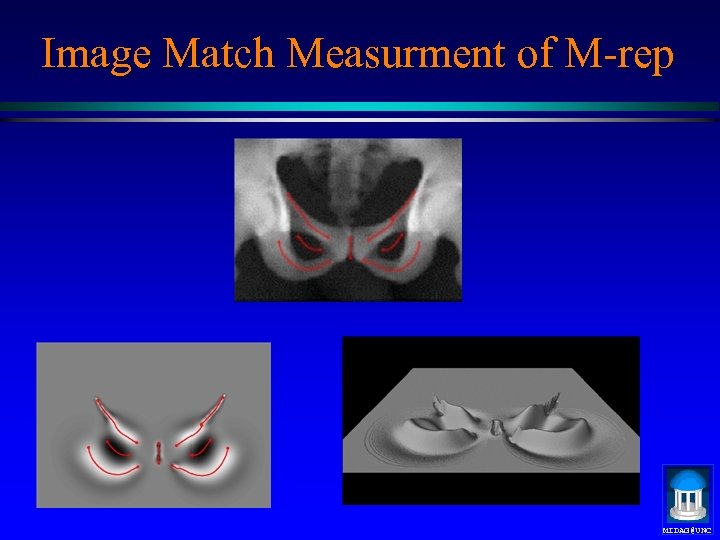
Image Match Measurment of M-rep MIDAG@UNC
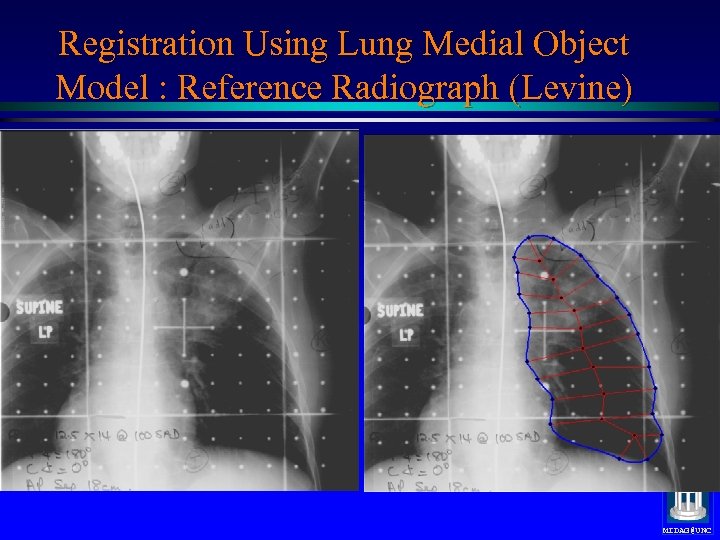
Registration Using Lung Medial Object Model : Reference Radiograph (Levine) Medial nets, positions only MIDAG@UNC
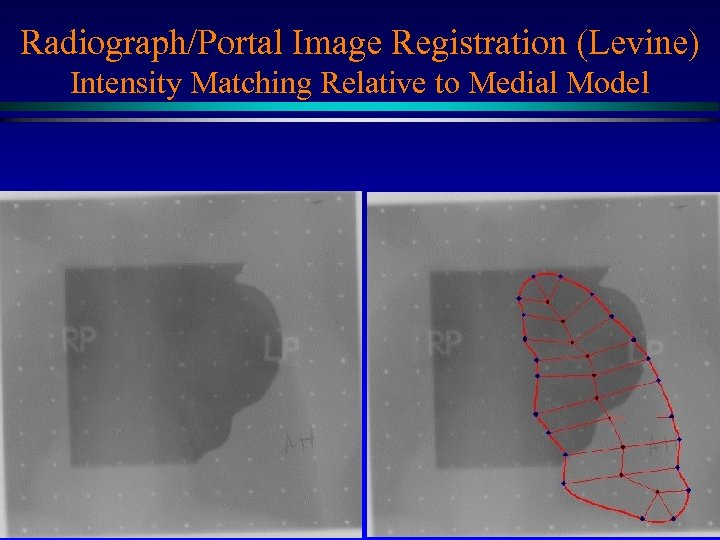
Radiograph/Portal Image Registration (Levine) Intensity Matching Relative to Medial Model Medial net MIDAG@UNC
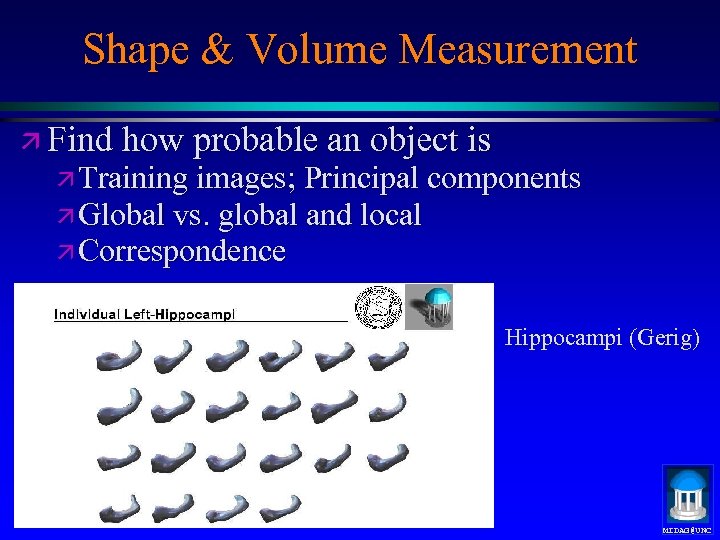
Shape & Volume Measurement ä Find how probable an object is ä Training images; Principal components ä Global vs. global and local ä Correspondence Hippocampi (Gerig) MIDAG@UNC

Modes of Global Deformation Training set: Mode 1: x = xmean + b 1 p 1 Mode 2: x = xmean + b 2 p 2 Mode 3: x = xmean + b 3 p 3 MIDAG@UNC
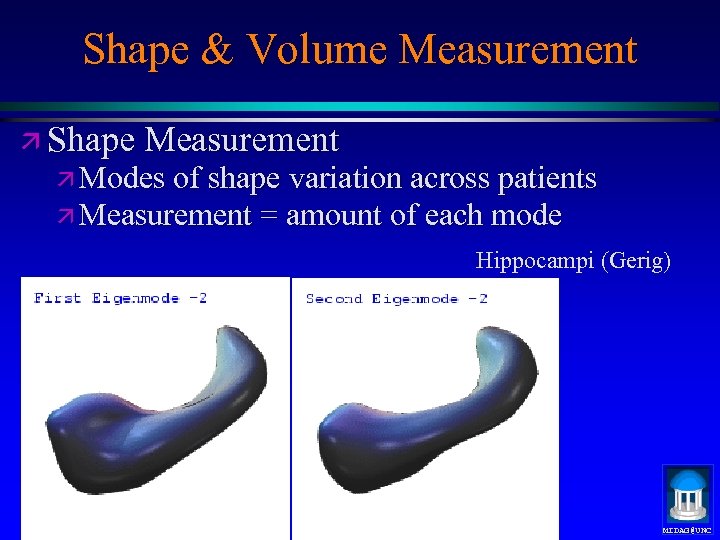
Shape & Volume Measurement ä Shape Measurement ä Modes of shape variation across patients ä Measurement = amount of each mode Hippocampi (Gerig) MIDAG@UNC
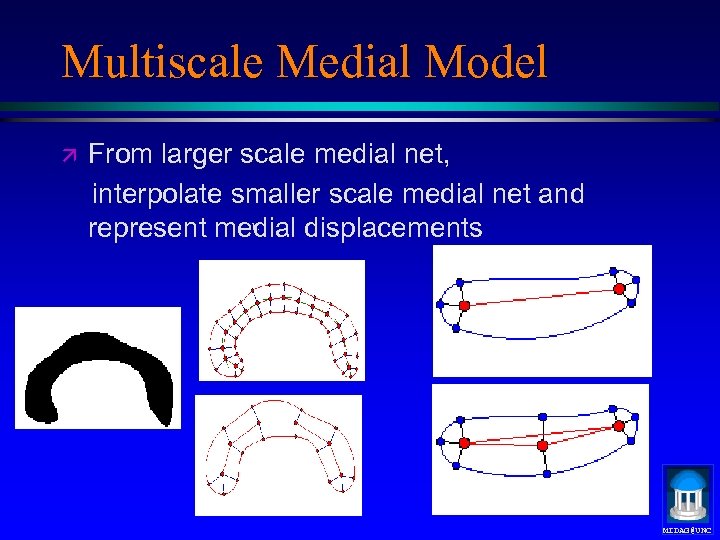
Multiscale Medial Model ä From larger scale medial net, interpolate smaller scale medial net and represent medial displacements b. MIDAG@UNC

Summary: What shape representation is for in medicine ä Analysis from images ä Extract the “anatomic object”-shaped object ä Register based on the objects ä Diagnose based on shape and volume ä Medical science via shape ä Shape and biology ä Shape-based diagnostic approaches ä Shape-based therapy planning and delivery approaches MIDAG@UNC

Shape Sciences ä Medicine ä Biology ä Geometry ä Statistics ä Image Analysis ä Computer Graphics MIDAG@UNC

The End MIDAG@UNC

Options for Primitives ä Space: xi for grid elements ä Landmarks: xi described by local geometry ä Boundary: (xi , normali) spaced along boundary ä Figural: nets of diatoms sampling figures MIDAG@UNC
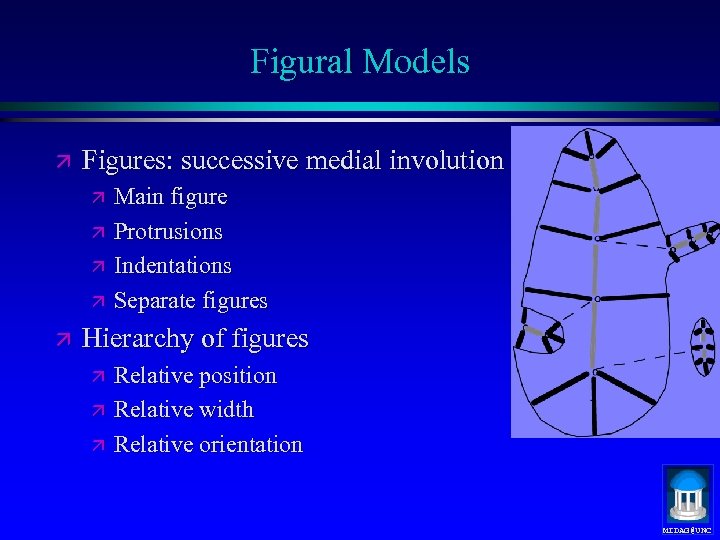
Figural Models ä Figures: successive medial involution ä ä ä Main figure Protrusions Indentations Separate figures Hierarchy of figures ä ä ä Relative position Relative width Relative orientation MIDAG@UNC
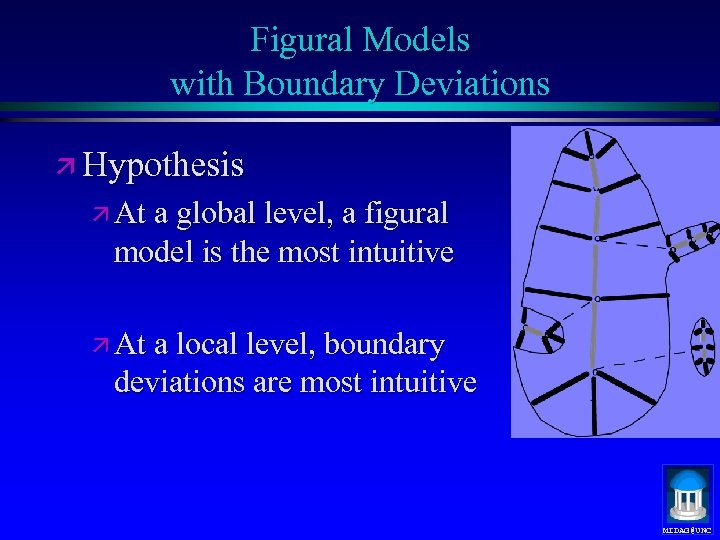
Figural Models with Boundary Deviations ä Hypothesis ä At a global level, a figural model is the most intuitive ä At a local level, boundary deviations are most intuitive MIDAG@UNC
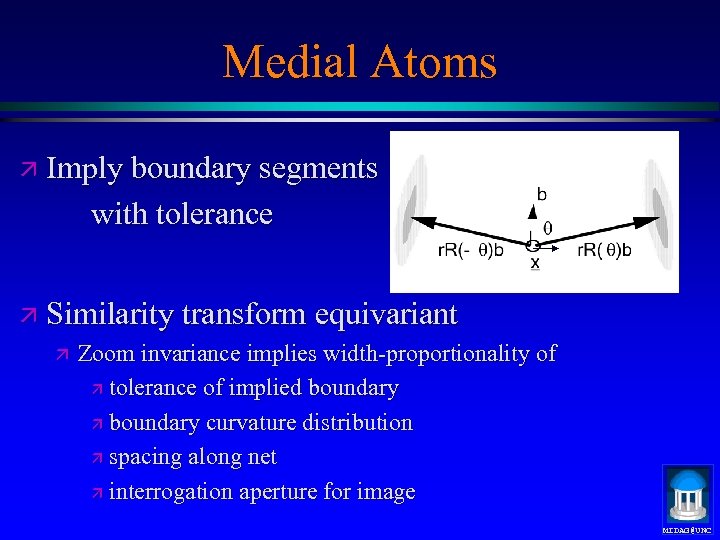
Medial Atoms ä Imply boundary segments with tolerance ä Similarity transform equivariant ä Zoom invariance implies width-proportionality of ä tolerance of implied boundary ä boundary curvature distribution ä spacing along net ä interrogation aperture for image MIDAG@UNC

Need for Special End Primitives ä Represent ä non-blobby objects ä angulated edges, corners, creases ä still allow rounded edges , corners, creases ä allow bent edges ä But ä Avoid infinitely fine medial sampling ä Maintain tangency, symmetry principles MIDAG@UNC

Coarse-to-fine representation ä For each of three levels ä Figural hierarchy ä For each figure, net chain, successively smaller tolerance ä For each net tile, boundary displacement chain MIDAG@UNC
Multiscale Medial Model ä From larger scale medial net ä Coarsely sampled ä Smooother figurally implied boundary ä Larger tolerance ä Interpolate smaller scale medial net ä Finer sampled ä More detail in figurally implied boundary ä Smaller tolerance ä Represent medial displacements MIDAG@UNC
Multiscale Medial/Boundary Model ä From medial net ä Coarsely sampled, smoother implied boundary ä Larger tolerance ä Represent boundary displacements along implied normals ä Finer sampled, more detail in boundary ä Smaller tolerance MIDAG@UNC
Shape Repres’n in Image Analysis ä Segmentation äFind the most probable deformed mean model, given the image äProbability involves ä Probability of the deformed model ä Probability of the image, given the deformed model MIDAG@UNC
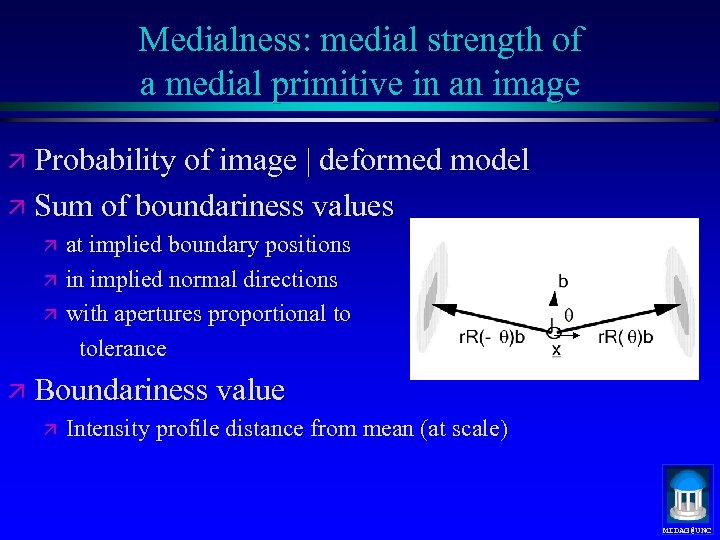
Medialness: medial strength of a medial primitive in an image ä Probability of image | deformed model ä Sum of boundariness values ä at implied boundary positions ä in implied normal directions ä with apertures proportional to tolerance ä Boundariness value ä Intensity profile distance from mean (at scale) MIDAG@UNC
Shape Rep’n in Image Analysis ä Segmentation ä Find the most probable deformed mean model, given the image ä Registration ä Find the most probable deformation, given the image ä Shape Measurement ä Find how probable a deformed model is MIDAG@UNC

Object Shape Representations for Medicine to Manufacturing ä Figural models, at successive levels of tolerance ä Boundary displacements ä Work in progress ä Segmentation and registration tools ä Statistical analysis of object populations ä CAD tools, incl. direct rendering ä… MIDAG@UNC
d199feb5c77224ea2a63d413de5998e2.ppt