1c5515f7f4238e00a7fc621a7ef62f27.ppt
- Количество слайдов: 48

The TMAJ Software Project and Database: Angelo M. De Marzo MD Ph. D James Morgan BS November 12, 2007
Introduction n Many putative new disease target genes with diagnostic, prognostic, and therapeutic applications Validation requires many samples Quantitative RT-PCR or protein arrays have disadvantages n n n Genes may be expressed in multiple different cell types In situ analyses: ideal but generally slow Tissue microarrays address some of these problems
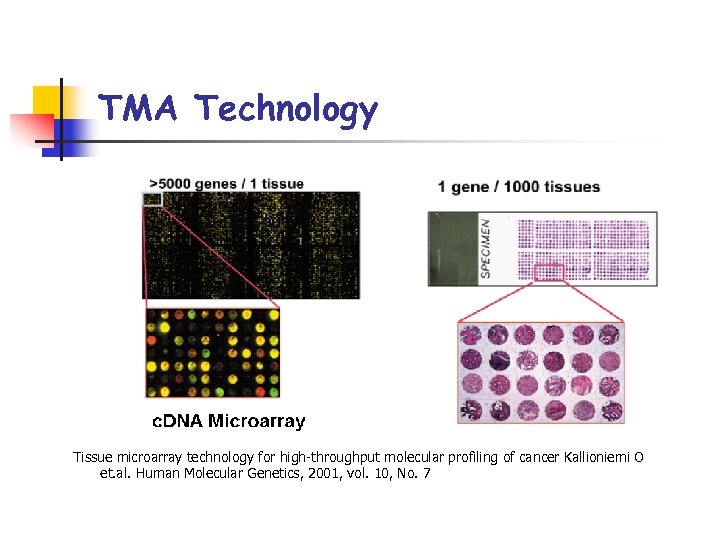
TMA Technology Tissue microarray technology for high-throughput molecular profiling of cancer Kallioniemi O et. al. Human Molecular Genetics, 2001, vol. 10, No. 7
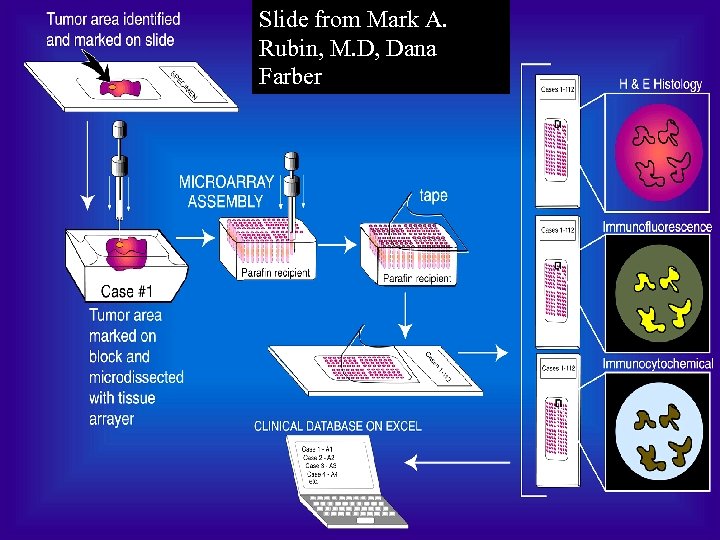
Slide from Mark A. Rubin, M. D, Dana Farber
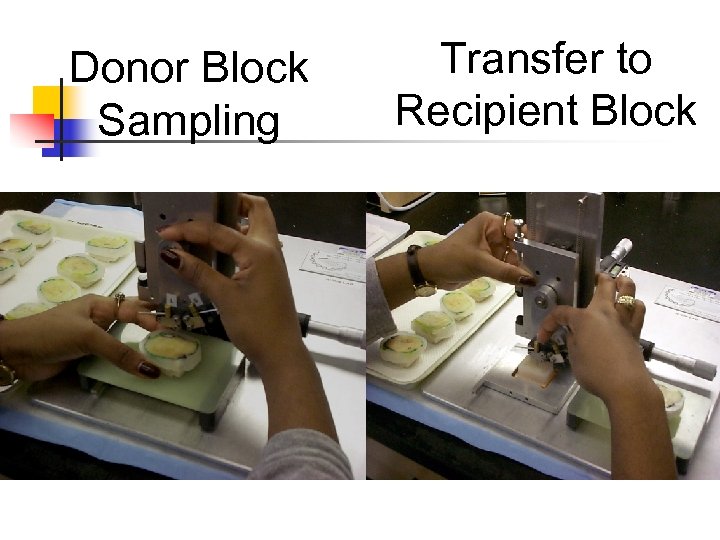
Donor Block Sampling Transfer to Recipient Block
Tissue Microarray Advantages § § § High throughput Expands tissue use Uniform reaction conditions Built-in controls Economize use of reagents Facilitates data recording and linking to clinical data
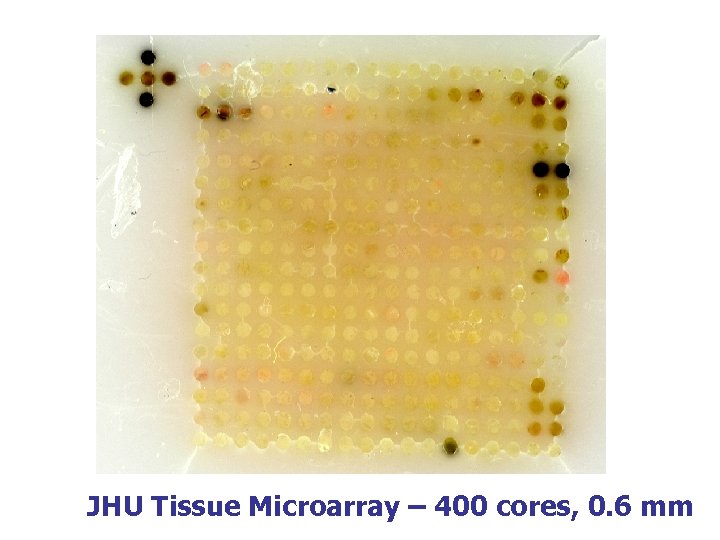
JHU Tissue Microarray – 400 cores, 0. 6 mm
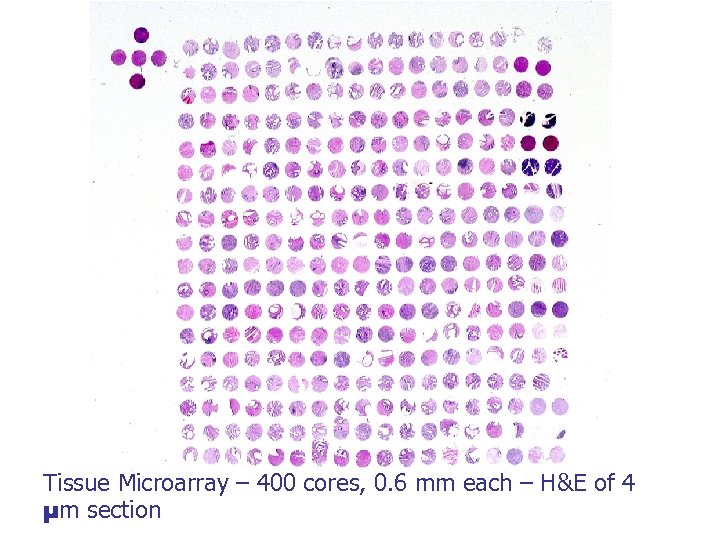
Tissue Microarray – 400 cores, 0. 6 mm each – H&E of 4 µm section

Digital Image Acquisition § § Can use conventional microscopes Record data in spreadsheet: diagnoses and interpretations Or the data can be recorded on paper for later entry into a spreadsheet or database Major Problem: § Easy to loose track of the x and y coordinates of given spots
Tissue Microarray Image Acquisition Aperio Scan. Scope ACIS II, DAKO

Need for Data Management 200 TMAs from Johns Hopkins TMA lab

What is TMAJ? n TMA-J is a set of open source software tools and backend database structure to facilitate management and analysis of tissue microarrays and associated pathology and image data
What Does TMAJ Do? The software applications provide a platform for: n n n Entering pathology data Managing users and permissions Designing TMAs Viewing and scoring TMA (and other) images online Side-by-side viewing of serial TMA images from slides stained for different biomarkers Publishing large numbers of TMA images and datasets on the Internet
What Does TMAJ Do? The Database Tracts: n Clinical information about patients n Pathology specimens and associated data n Pathology tissue blocks n Tissue Microarray cores n TMA Blocks n TMA Slides n TMA core images n TMA image scoring data: manual or semiautomated

Primary Goals of System n n n n Address security issues Remove or isolate patient identifiers Manage multiple organ systems Develop web based interface Scalable to accommodate large number of simultaneous users Storage of large sets of images with diagnoses Data structure compatible with emerging standards for easy data exchange n n Ca. BIG compatibility (to be defined) The tissue microarray data exchange specification: n Berman et al. , (http: //www. pubmedcentral. nih. gov/articlerender. fcgi ? artid=165444)

Database Design
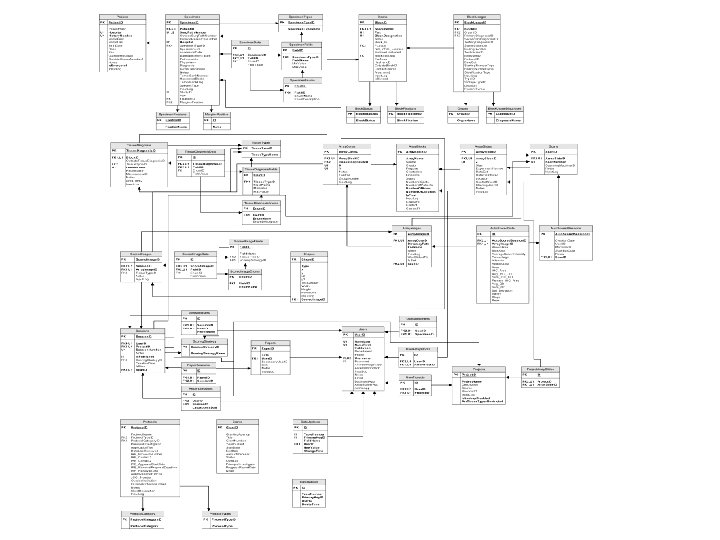
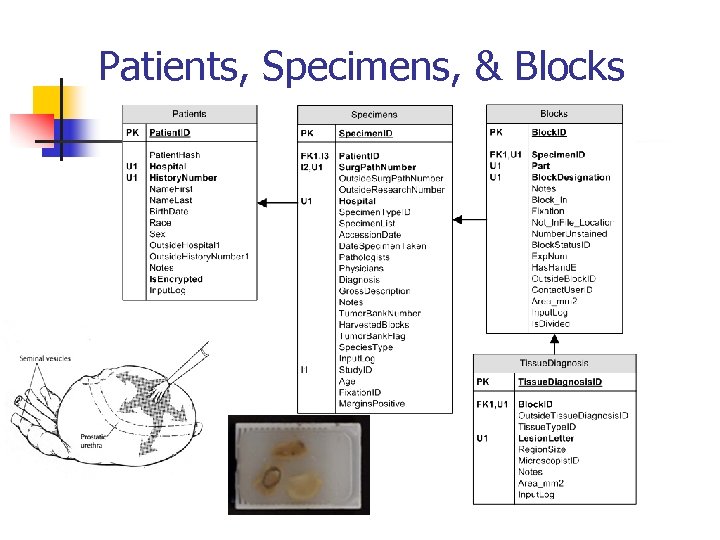
Patients, Specimens, & Blocks The Patients, Specimens, Blocks, and Tissue Diagnosis tables all form a one-to-many relationship.
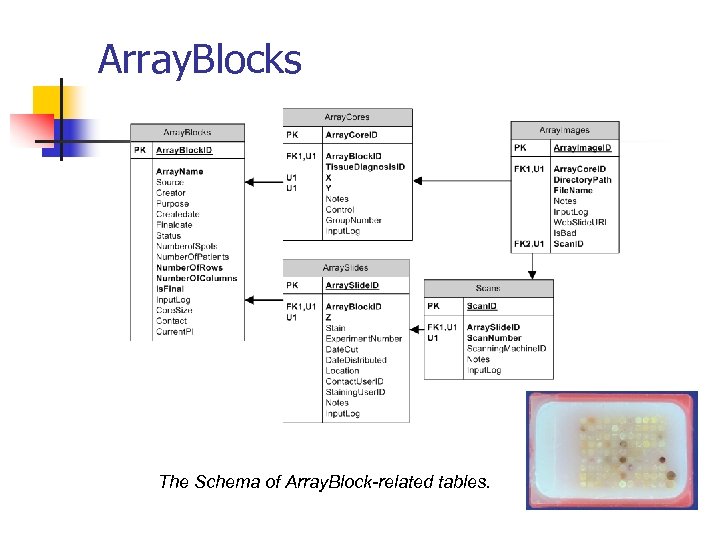
Array. Blocks The Schema of Array. Block-related tables.

Security: Protecting Patient Information n n Database stored on a secure server Identifiable patient information in encrypted tables (Approved by the IRB) Researchers have no access to patient identifiers Creates virtual separate entities: “clinical database” and “research database”
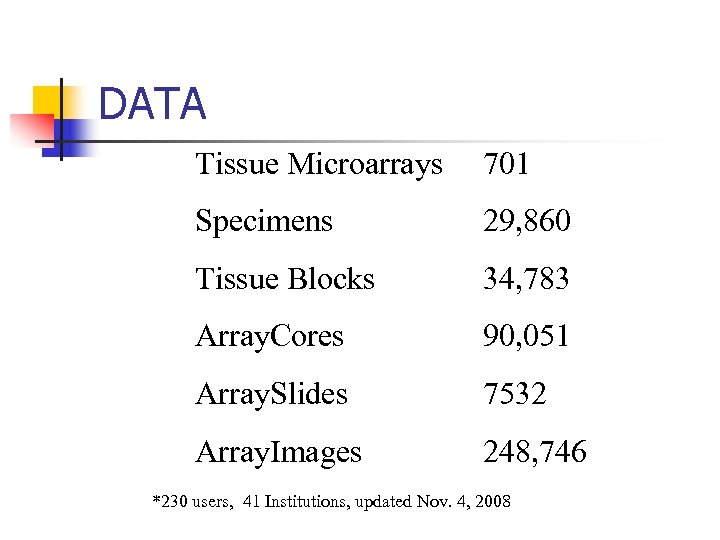
DATA Tissue Microarrays 701 Specimens 29, 860 Tissue Blocks 34, 783 Array. Cores 90, 051 Array. Slides 7532 Array. Images 248, 746 *230 users, 41 Institutions, updated Nov. 4, 2008
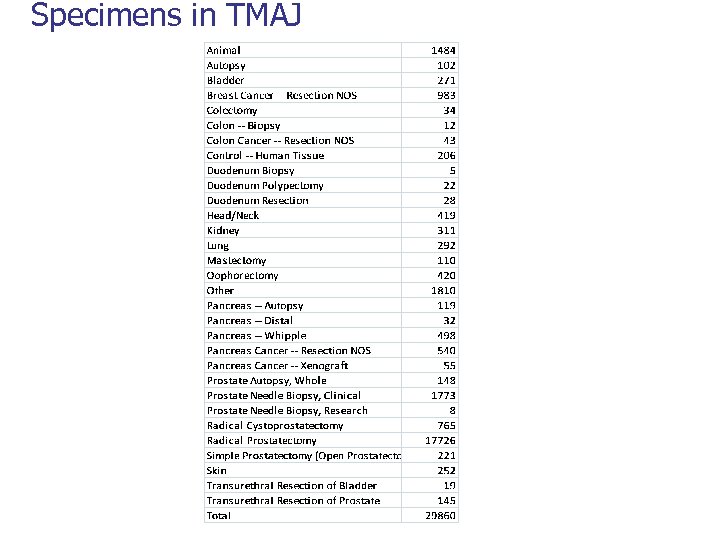
Specimens in TMAJ
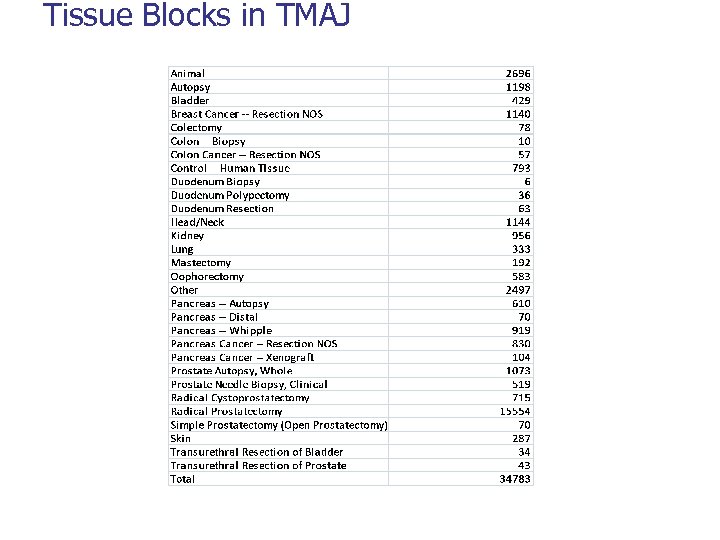
Tissue Blocks in TMAJ

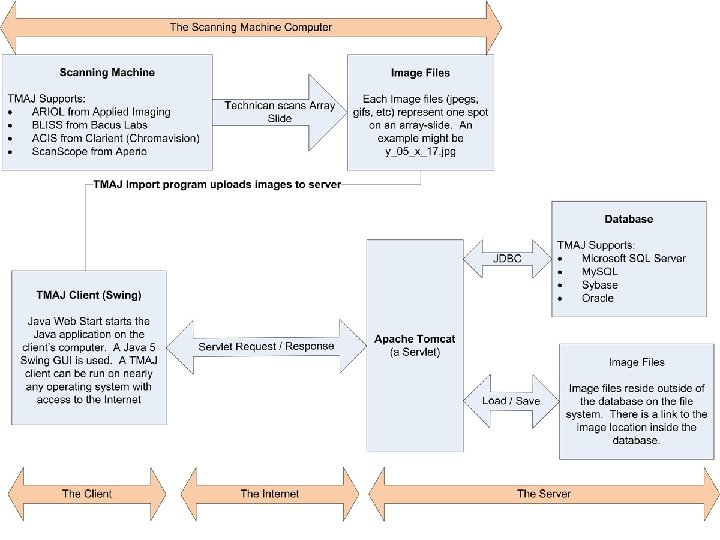
Applications – Java from Sun Microsystems Java Web Start Software n n Java Web Start software provides a browserindependent architecture for deploying Java technology-based applications to the client desktop Each application runs on a dedicated Java Virtual Machine (JVM)

Applications & Screenshots
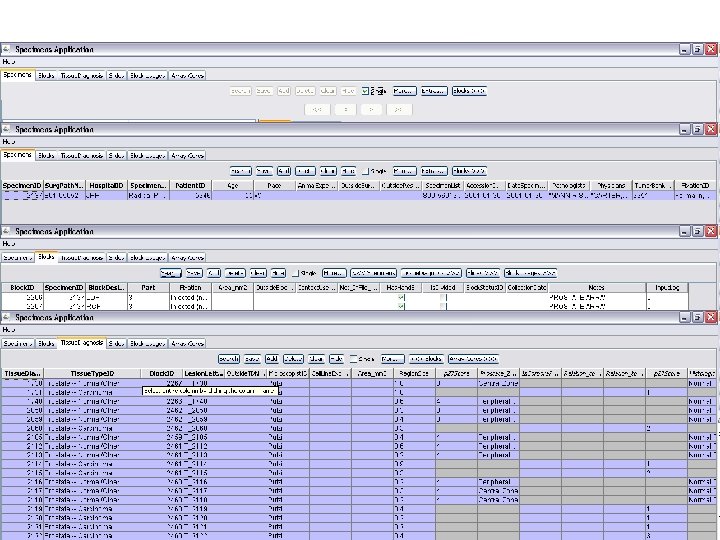
Specimens Application n This application allows for detailed input of data on individual specimens and donor-tissue-blocks.
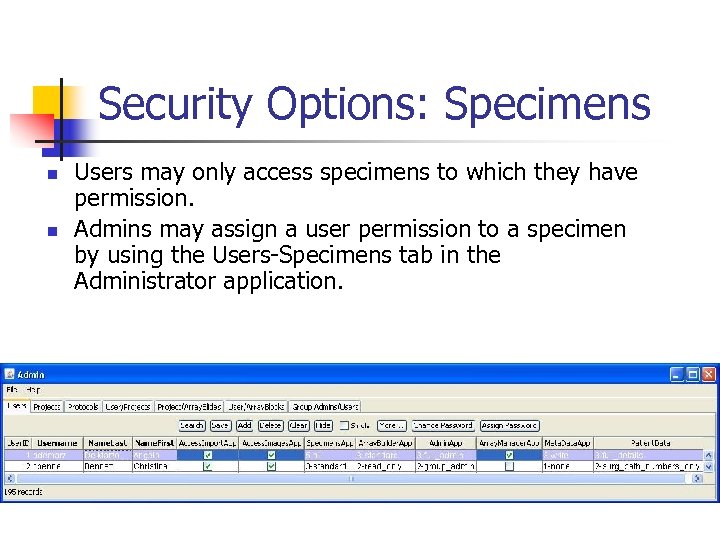
Security Options: Specimens n n Users may only access specimens to which they have permission. Admins may assign a user permission to a specimen by using the Users-Specimens tab in the Administrator application.
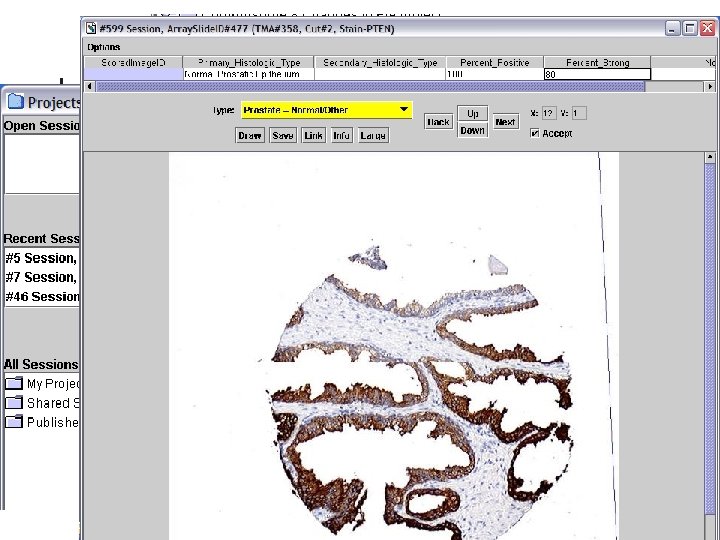
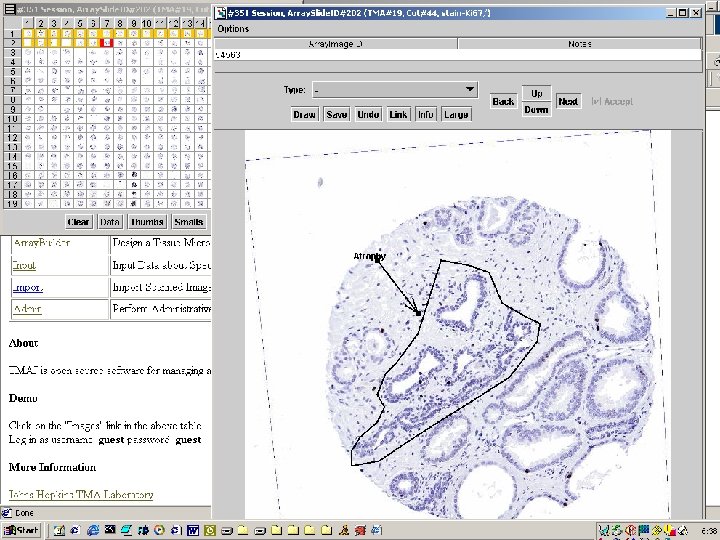
Images Application
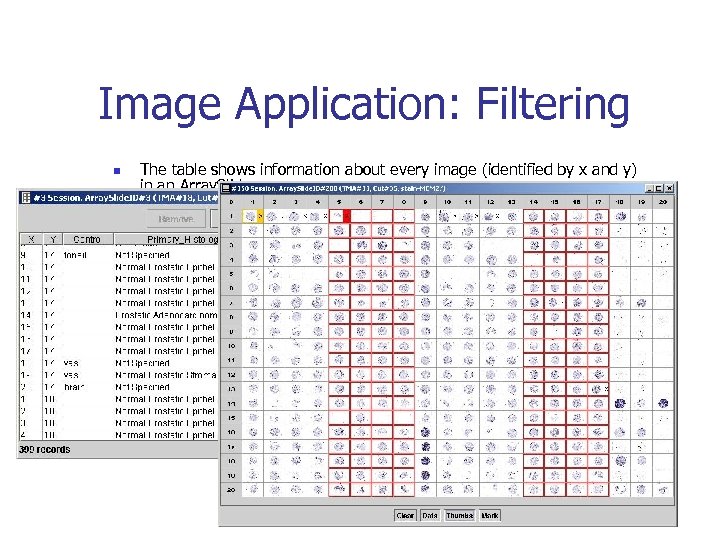
Image Application: Filtering n n The table shows information about every image (identified by x and y) in an Array. Slide. Images identified as “Prostate – Carcinoma” are highlighted in red.
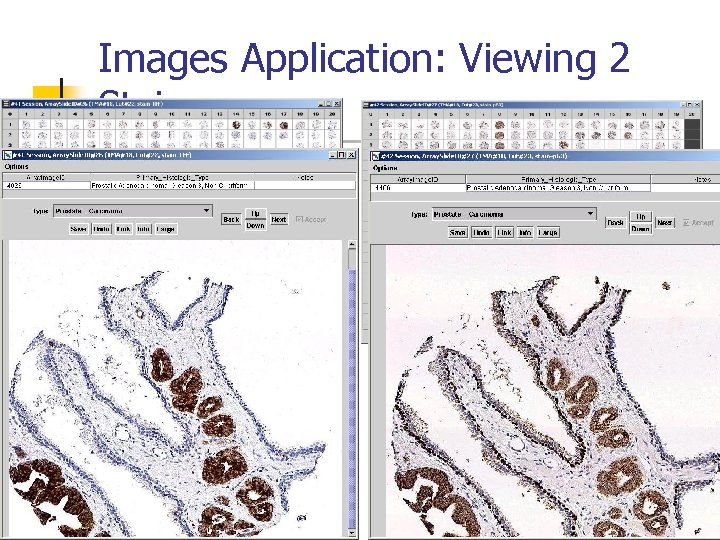
Images Application: Viewing 2 Stains
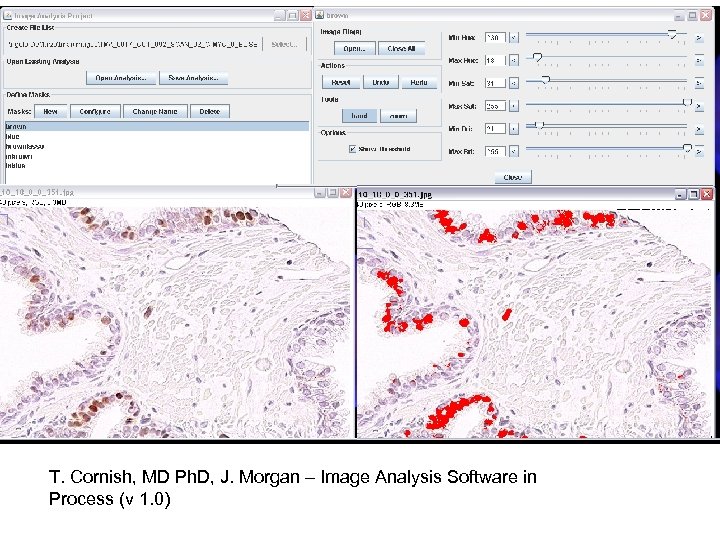
T. Cornish, MD Ph. D, J. Morgan – Image Analysis Software in Process (v 1. 0)

Publishing TMA Images and Scoring Data Over the Internet n n Roughly modeled after Stanford Microarray Database Concept: Once a study is published by a journal, all TMA diagnoses, image, scoring and non-protected clinical data can be “published” as supplemental data to the Internet for public online viewing or down loading n TMAJ Images now linked to “Proteinpedia” database (http: //humanproteinpedia. org) by Akhilesh Pandy, MD Ph. D. n

For More Information n n http: //tmaj. pathology. jhmi. edu To see published images n login to tmaj as a guest and then click the Images button. n n Username: guest Password: guest

Institutions Using TMAJ n n n Johns Hopkins University Harvard Dana Farber Cancer Institute Cleveland Clinic University of Texas Southwestern Vanderbilt University

Dynamic Fields in TMAJ What are Dynamic Fields, why are they important, and how are they managed in TMAJ?
Dynamic Fields n n n Different organ systems will have different recorded data. For example the Gleason score is only relevant to the prostate. Dynamic fields allow TMAJ to keep track of different data for different organ systems. TMAJ can have dynamic fields added at any time through the GUI. Database access is not needed and the code does not need to be recompiled.
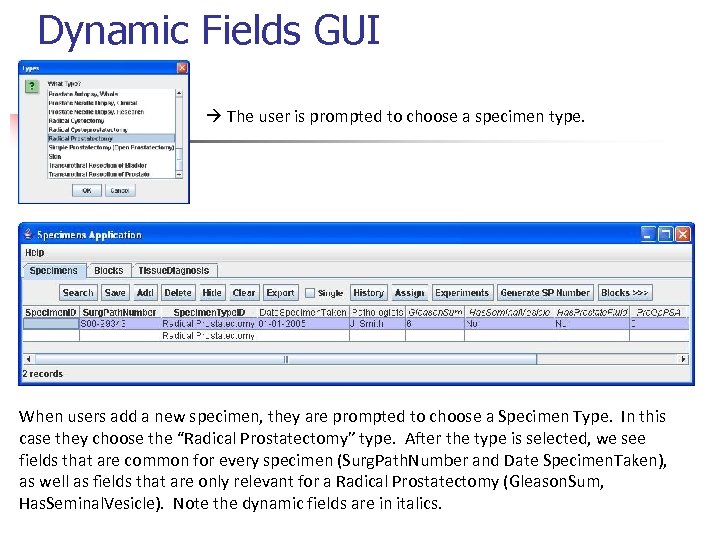
Dynamic Fields GUI The user is prompted to choose a specimen type. When users add a new specimen, they are prompted to choose a Specimen Type. In this case they choose the “Radical Prostatectomy” type. After the type is selected, we see fields that are common for every specimen (Surg. Path. Number and Date Specimen. Taken), as well as fields that are only relevant for a Radical Prostatectomy (Gleason. Sum, Has. Seminal. Vesicle). Note the dynamic fields are in italics.
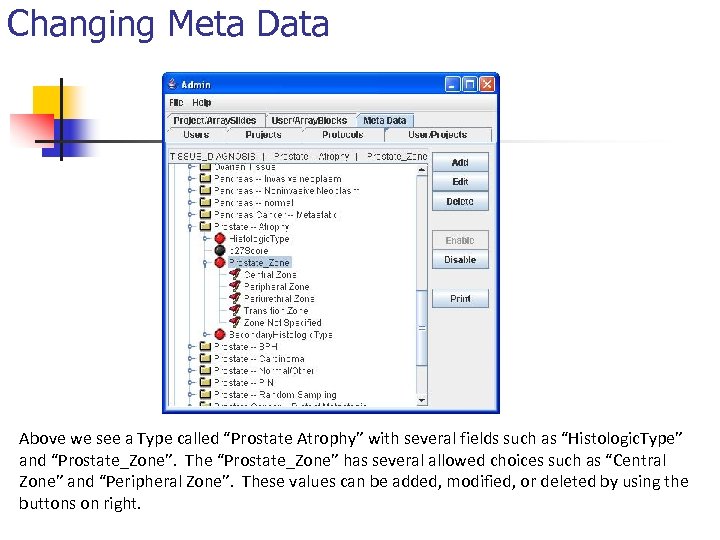
Changing Meta Data Above we see a Type called “Prostate Atrophy” with several fields such as “Histologic. Type” and “Prostate_Zone”. The “Prostate_Zone” has several allowed choices such as “Central Zone” and “Peripheral Zone”. These values can be added, modified, or deleted by using the buttons on right.

One Approach: A Key-Values Table n n A Key Values table would only have 3 fields: A Key (such a Prostate Weight), a value, and a foreign key that links the record back to the main table (such as the Specimens table). We did not use this approach because it does not keep track of the meta-data. Meta-Data is data that describes data, and in this case it would be the type (Prostate), the field for the type (Prostate Weight), and any allowed choices.
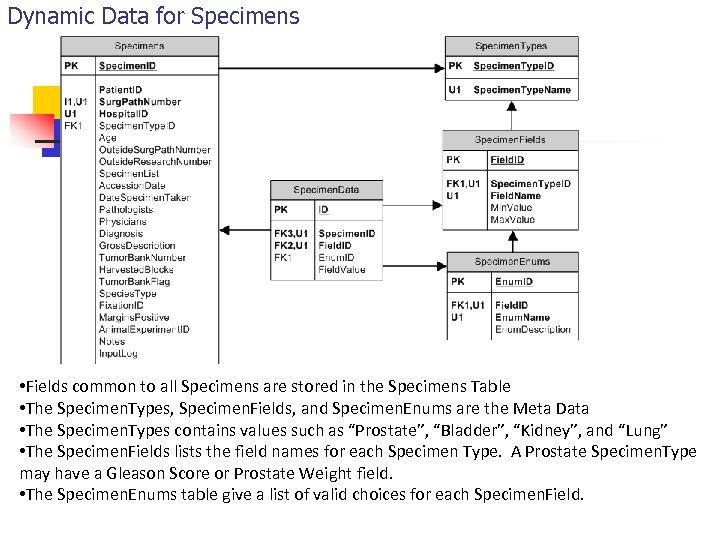
Dynamic Data for Specimens • Fields common to all Specimens are stored in the Specimens Table • The Specimen. Types, Specimen. Fields, and Specimen. Enums are the Meta Data • The Specimen. Types contains values such as “Prostate”, “Bladder”, “Kidney”, and “Lung” • The Specimen. Fields lists the field names for each Specimen Type. A Prostate Specimen. Type may have a Gleason Score or Prostate Weight field. • The Specimen. Enums table give a list of valid choices for each Specimen. Field.
TMAJ & Frida Integration n The image analysis software package Frida has been integrated with TMAJ
Using Image Analysis in TMAJ n New Image Analysis Sessions are created for a scanned arrayslide
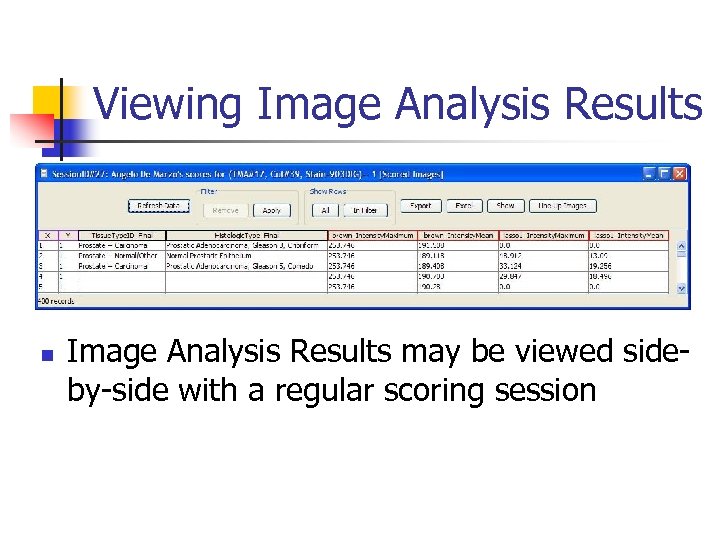
Viewing Image Analysis Results n Image Analysis Results may be viewed sideby-side with a regular scoring session

Acknowledgements Tissue Microarray Lab Marc Halushka MD Ph. D Helen Fedor BS Marcella Southerland BS Qizhi Zheng MD James Morgan BS Kristen Lecksell BS De Marzo Lab Jessica Hicks BS Toby Cornish MD Ph. D
1c5515f7f4238e00a7fc621a7ef62f27.ppt