2a342ee629e578a29bd4c84b0bffee20.ppt
- Количество слайдов: 43
 The Role of Ontologies in Improved Scholarly Communication Philip E. Bourne University of California San Diego pbourne@ucsd. edu http: //www. sdsc. edu/pb
The Role of Ontologies in Improved Scholarly Communication Philip E. Bourne University of California San Diego pbourne@ucsd. edu http: //www. sdsc. edu/pb
 My Perspective … • Ontology Developer (years ago – mm. CIF - Bioinformatics 2002 18: 1280 -128) • Database Developer – RCSB PDB • Supporter of open access (provided there is a business model) - editor in chief of PLo. S Computational Biology • Co-founder - Sci. Vee Inc. • I am becoming increasingly interested in scholarly communication • I use ontologies to support this work
My Perspective … • Ontology Developer (years ago – mm. CIF - Bioinformatics 2002 18: 1280 -128) • Database Developer – RCSB PDB • Supporter of open access (provided there is a business model) - editor in chief of PLo. S Computational Biology • Co-founder - Sci. Vee Inc. • I am becoming increasingly interested in scholarly communication • I use ontologies to support this work
 Objective Today • Describe how we are using ontologies to try and improve scholarly communication • Motivate you towards thinking about ontologies that should be developed • Learn from you where we might spend our efforts
Objective Today • Describe how we are using ontologies to try and improve scholarly communication • Motivate you towards thinking about ontologies that should be developed • Learn from you where we might spend our efforts
 First Consider What Motivates Us to Improve Scholarly Communication
First Consider What Motivates Us to Improve Scholarly Communication
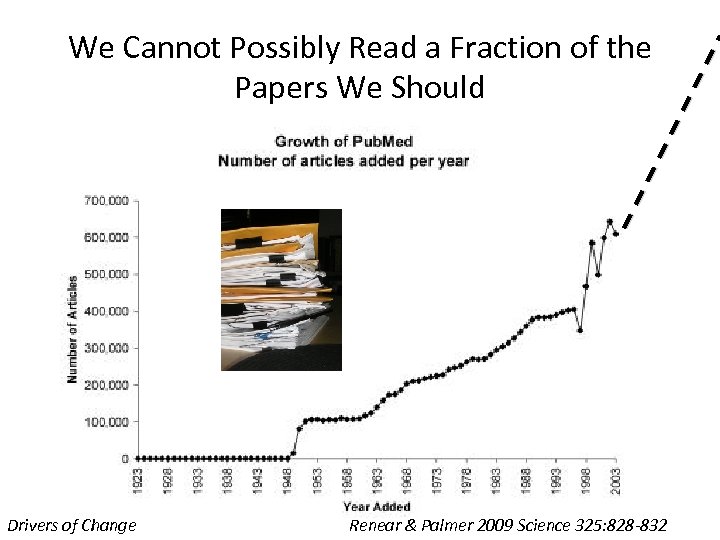 We Cannot Possibly Read a Fraction of the Papers We Should Drivers of Change Renear & Palmer 2009 Science 325: 828 -832
We Cannot Possibly Read a Fraction of the Papers We Should Drivers of Change Renear & Palmer 2009 Science 325: 828 -832
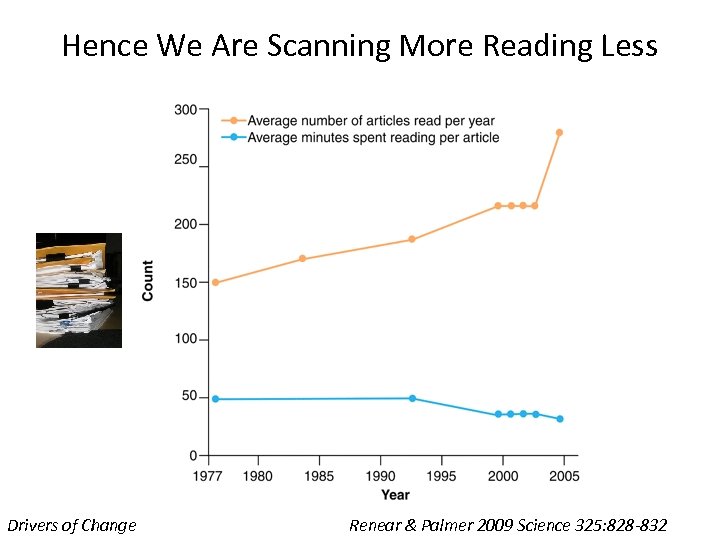 Hence We Are Scanning More Reading Less Drivers of Change Renear & Palmer 2009 Science 325: 828 -832
Hence We Are Scanning More Reading Less Drivers of Change Renear & Palmer 2009 Science 325: 828 -832
 The Truth About the Scientific e. Laboratory • I have ? ? mail folders! • The intellectual memory of my laboratory is in those folders • This is an unhealthy hub and spoke mentality Drivers of Change
The Truth About the Scientific e. Laboratory • I have ? ? mail folders! • The intellectual memory of my laboratory is in those folders • This is an unhealthy hub and spoke mentality Drivers of Change
 The Truth About the Scientific e. Laboratory • I generate way more negative that positive data, but where is it? • Content management is a mess – Slides, posters…. . – Data, lab notebooks …. – Collaborations, Journal clubs … • Software is open but where is it? • Farewell is for the data too Drivers of Change Computational Biology Resources Lack Persistence and Usability. PLo. S Comp. Biol. 4(7): e 1000136
The Truth About the Scientific e. Laboratory • I generate way more negative that positive data, but where is it? • Content management is a mess – Slides, posters…. . – Data, lab notebooks …. – Collaborations, Journal clubs … • Software is open but where is it? • Farewell is for the data too Drivers of Change Computational Biology Resources Lack Persistence and Usability. PLo. S Comp. Biol. 4(7): e 1000136
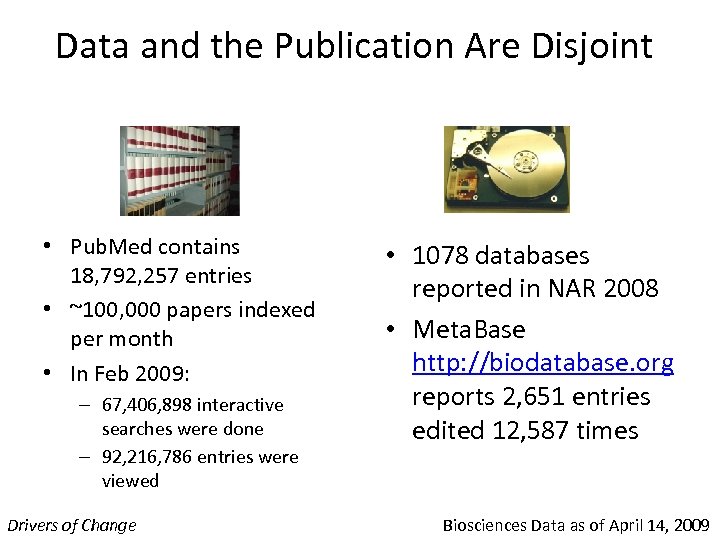 Data and the Publication Are Disjoint • Pub. Med contains 18, 792, 257 entries • ~100, 000 papers indexed per month • In Feb 2009: – 67, 406, 898 interactive searches were done – 92, 216, 786 entries were viewed Drivers of Change • 1078 databases reported in NAR 2008 • Meta. Base http: //biodatabase. org reports 2, 651 entries edited 12, 587 times Biosciences Data as of April 14, 2009
Data and the Publication Are Disjoint • Pub. Med contains 18, 792, 257 entries • ~100, 000 papers indexed per month • In Feb 2009: – 67, 406, 898 interactive searches were done – 92, 216, 786 entries were viewed Drivers of Change • 1078 databases reported in NAR 2008 • Meta. Base http: //biodatabase. org reports 2, 651 entries edited 12, 587 times Biosciences Data as of April 14, 2009
 Publishing Limitations • A paper is an artifact of a previous era • It is not the logical end product of e. Science, hence: – Work is omitted – Article vs supplement is a mess – Visualization may be limited – Interaction and enquiry are non-existent – Rich media can help, but are rarely used Drivers of Change
Publishing Limitations • A paper is an artifact of a previous era • It is not the logical end product of e. Science, hence: – Work is omitted – Article vs supplement is a mess – Visualization may be limited – Interaction and enquiry are non-existent – Rich media can help, but are rarely used Drivers of Change
 We Need to do Better & The Game is Afoot It is being driven from the top down and the bottom up
We Need to do Better & The Game is Afoot It is being driven from the top down and the bottom up
 Ontologies & Semantic Tagging
Ontologies & Semantic Tagging
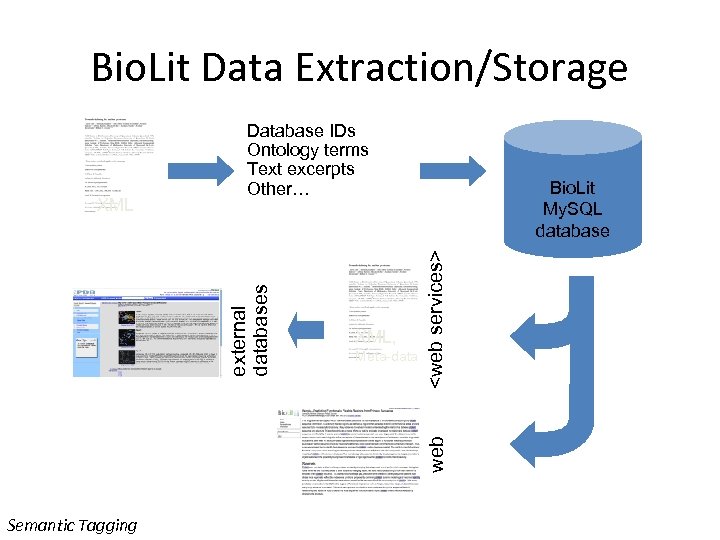 Bio. Lit Data Extraction/Storage Meta-data
Bio. Lit Data Extraction/Storage Meta-data
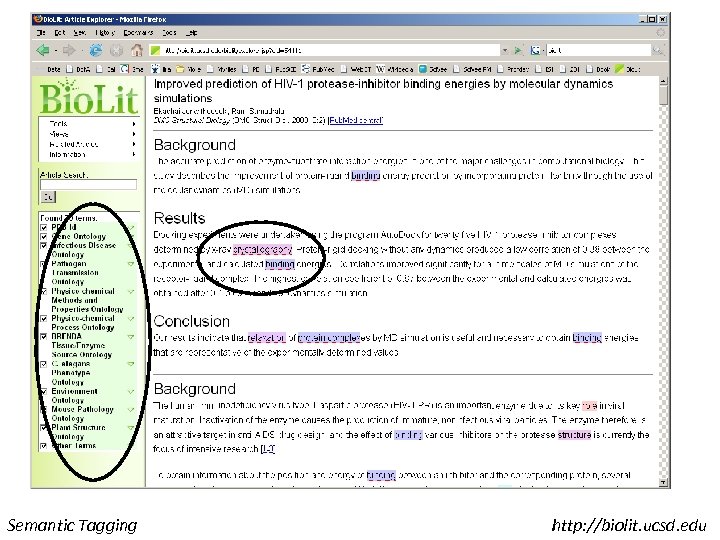
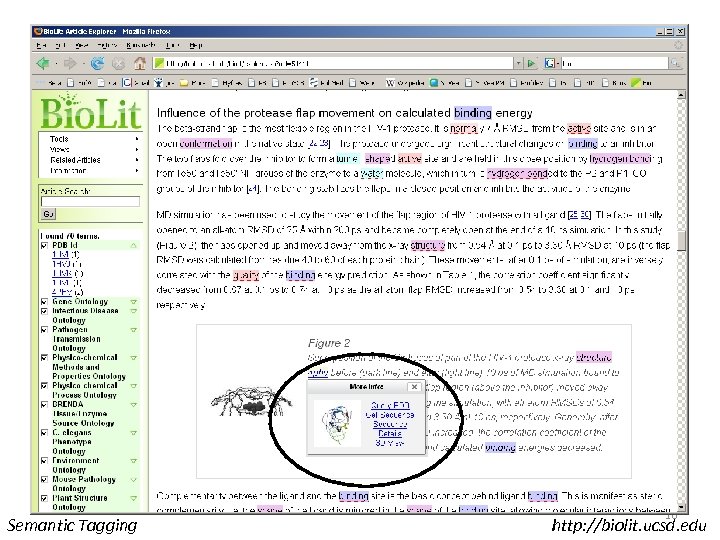
 Tagging of Pub. Med Central • Ontologies read from OBO Files • Words converted to tree structures • Matched to every non-trivial word in the paper • Matches tagged • A long paper can be matched to GO in less than 30 seconds Semantic Tagging http: //biolit. ucsd. edu
Tagging of Pub. Med Central • Ontologies read from OBO Files • Words converted to tree structures • Matched to every non-trivial word in the paper • Matches tagged • A long paper can be matched to GO in less than 30 seconds Semantic Tagging http: //biolit. ucsd. edu
 Semantic Tagging http: //biolit. ucsd. edu
Semantic Tagging http: //biolit. ucsd. edu
 ICTP Trieste, December 10, 2007 Semantic Tagging 16 http: //biolit. ucsd. edu
ICTP Trieste, December 10, 2007 Semantic Tagging 16 http: //biolit. ucsd. edu
 Provision of Webservices to this tagging may be the most valuable contribution. . Semantic Tagging
Provision of Webservices to this tagging may be the most valuable contribution. . Semantic Tagging
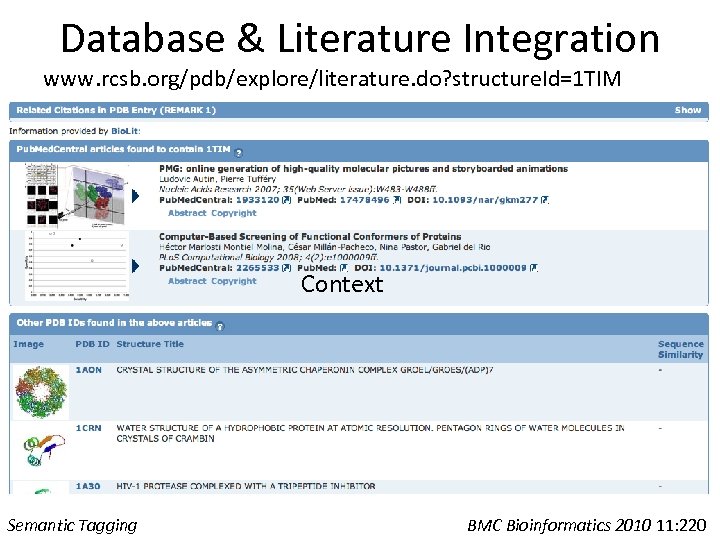 Database & Literature Integration www. rcsb. org/pdb/explore/literature. do? structure. Id=1 TIM Context Semantic Tagging BMC Bioinformatics 2010 11: 220
Database & Literature Integration www. rcsb. org/pdb/explore/literature. do? structure. Id=1 TIM Context Semantic Tagging BMC Bioinformatics 2010 11: 220
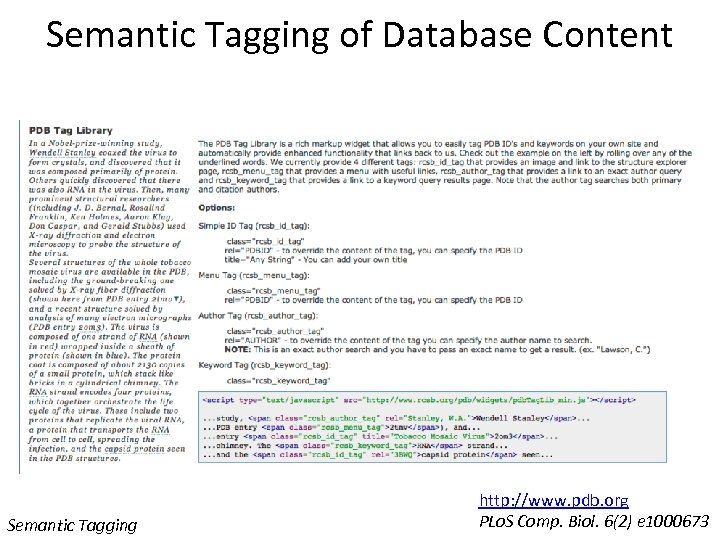 Semantic Tagging of Database Content Semantic Tagging http: //www. pdb. org PLo. S Comp. Biol. 6(2) e 1000673
Semantic Tagging of Database Content Semantic Tagging http: //www. pdb. org PLo. S Comp. Biol. 6(2) e 1000673
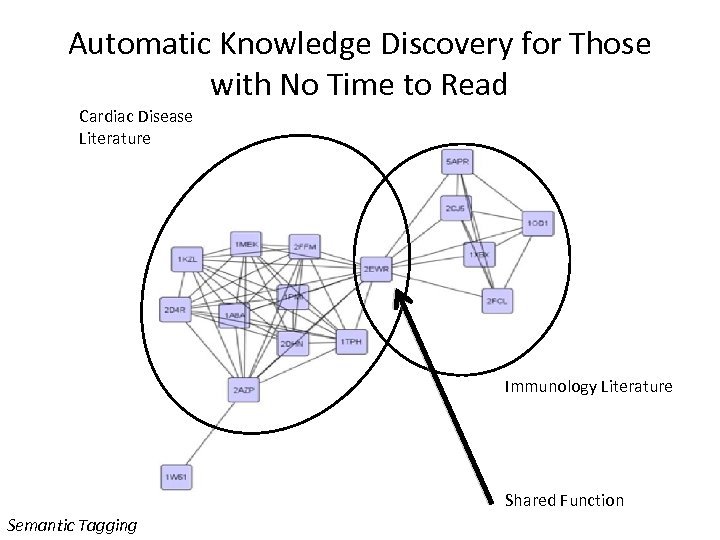 Automatic Knowledge Discovery for Those with No Time to Read Cardiac Disease Literature Immunology Literature Shared Function Semantic Tagging
Automatic Knowledge Discovery for Those with No Time to Read Cardiac Disease Literature Immunology Literature Shared Function Semantic Tagging
 This is Literature Post-processing Better to Get the Authors Involved • Authors are the absolute experts on the content • More effective distribution of labor • Add metadata before the article enters the publishing process Semantic Tagging BMC Bioinformatics 2010 11: 103
This is Literature Post-processing Better to Get the Authors Involved • Authors are the absolute experts on the content • More effective distribution of labor • Add metadata before the article enters the publishing process Semantic Tagging BMC Bioinformatics 2010 11: 103
 Word 2007 Add-in for Authors • Allows authors to add metadata as they write, before they submit the manuscript • Authors are assisted by automated term recognition – OBO ontologies – Database IDs • Metadata are embedded directly into the manuscript document via XML tags, OOXML format – Open – Machine-readable • Open source, Microsoft Public License Drivers of Change http: //www. codeplex. com/ucsdbiolit
Word 2007 Add-in for Authors • Allows authors to add metadata as they write, before they submit the manuscript • Authors are assisted by automated term recognition – OBO ontologies – Database IDs • Metadata are embedded directly into the manuscript document via XML tags, OOXML format – Open – Machine-readable • Open source, Microsoft Public License Drivers of Change http: //www. codeplex. com/ucsdbiolit
 Word 2007 Add-in Example of What it Looks Like - Ontologies • Inline Recognition, Highlighting, and Mark-up of Informative Terms – A recognized term will have a dotted, purple underline – Hovering generates a Smart Tag above the term • • add mark-up for this term ignore this term view the term in the ontology browser If a recognized term appears in more than one ontology, all instances of that term will be listed – Hovering over a marked-up term • option to apply mark-up to all recognized instances of term • stop recognizing a term – Pass ontology terms back to provider Semantic Tagging BMC Bioinformatics 2010 11: 103
Word 2007 Add-in Example of What it Looks Like - Ontologies • Inline Recognition, Highlighting, and Mark-up of Informative Terms – A recognized term will have a dotted, purple underline – Hovering generates a Smart Tag above the term • • add mark-up for this term ignore this term view the term in the ontology browser If a recognized term appears in more than one ontology, all instances of that term will be listed – Hovering over a marked-up term • option to apply mark-up to all recognized instances of term • stop recognizing a term – Pass ontology terms back to provider Semantic Tagging BMC Bioinformatics 2010 11: 103
 • Built-in Knowledge of Ontologies and Databases – Add-in provides a list of biomedical ontologies to download – and a list of databases for ID recognition (Gen. Bank/Ref. Seq, Uni. Prot, Protein Data Bank) – A user may also supply a URL to download other ontologies • Ontology Browser – allows a user to select an ontology and then navigate through it to view terms and their relationships BMC Bioinformatics 2010 11: 103
• Built-in Knowledge of Ontologies and Databases – Add-in provides a list of biomedical ontologies to download – and a list of databases for ID recognition (Gen. Bank/Ref. Seq, Uni. Prot, Protein Data Bank) – A user may also supply a URL to download other ontologies • Ontology Browser – allows a user to select an ontology and then navigate through it to view terms and their relationships BMC Bioinformatics 2010 11: 103
 Custom Metadata • Ontologies do not contain all usages of a concept • Add-in allows user to assign custom metadata • Human Disease Ontology term: Leukemia, T-Cell, HTLVII-Associated • Synonym: Atypical hairy cell leukemia (disorder) • Actual use in literature: – – hairy cell leukemia hairy-cell leukemia hairy T cell leukemia T cell hairy leukemia BMC Bioinformatics 2010 11: 103
Custom Metadata • Ontologies do not contain all usages of a concept • Add-in allows user to assign custom metadata • Human Disease Ontology term: Leukemia, T-Cell, HTLVII-Associated • Synonym: Atypical hairy cell leukemia (disorder) • Actual use in literature: – – hairy cell leukemia hairy-cell leukemia hairy T cell leukemia T cell hairy leukemia BMC Bioinformatics 2010 11: 103
 Synonym mapping, disambiguation • Inclusion of an additional set of synonyms for a term that reflect its use in natural language – Automated finding of synonyms in extant literature – Gather synonyms from term-mapping databases • Incorporate a more sophisticated term recognition approach into the add-in BMC Bioinformatics 2010 11: 103
Synonym mapping, disambiguation • Inclusion of an additional set of synonyms for a term that reflect its use in natural language – Automated finding of synonyms in extant literature – Gather synonyms from term-mapping databases • Incorporate a more sophisticated term recognition approach into the add-in BMC Bioinformatics 2010 11: 103
 Challenges • Author use – Familiarity with ontologies, terms – Agreement between co-authors • End-use of semantically enriched manuscript • Need to combine with NLM XML standard Semantic Tagging BMC Bioinformatics 2010 11: 103
Challenges • Author use – Familiarity with ontologies, terms – Agreement between co-authors • End-use of semantically enriched manuscript • Need to combine with NLM XML standard Semantic Tagging BMC Bioinformatics 2010 11: 103
 Challenges: Author Use IF one or more publishers fast tracked a paper that had semantic markup I would argue it would catch on in no time Semantic Tagging BMC Bioinformatics 2010 11: 103
Challenges: Author Use IF one or more publishers fast tracked a paper that had semantic markup I would argue it would catch on in no time Semantic Tagging BMC Bioinformatics 2010 11: 103
 Where we Need {Better} Ontologies 1. To Support Mashups Between Different Types of Scholarly Output
Where we Need {Better} Ontologies 1. To Support Mashups Between Different Types of Scholarly Output
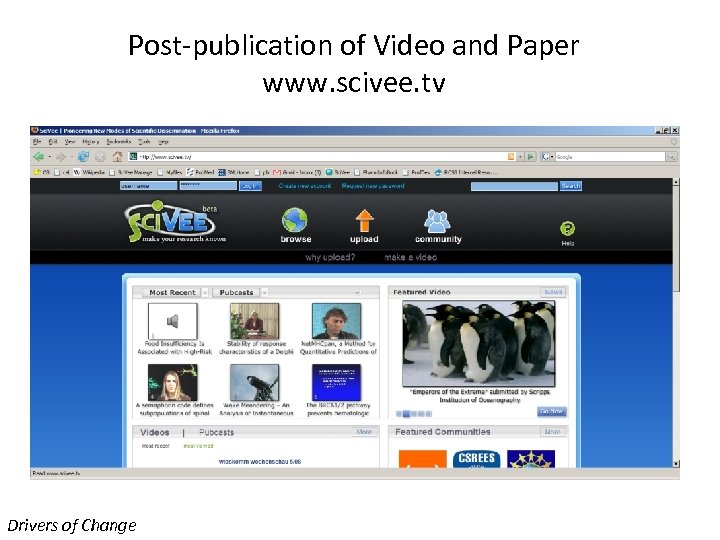 Post-publication of Video and Paper www. scivee. tv Drivers of Change
Post-publication of Video and Paper www. scivee. tv Drivers of Change
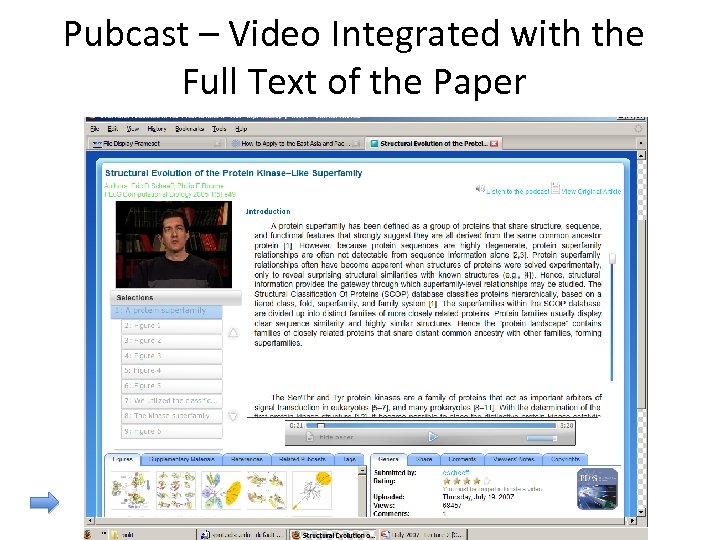 Pubcast – Video Integrated with the Full Text of the Paper
Pubcast – Video Integrated with the Full Text of the Paper
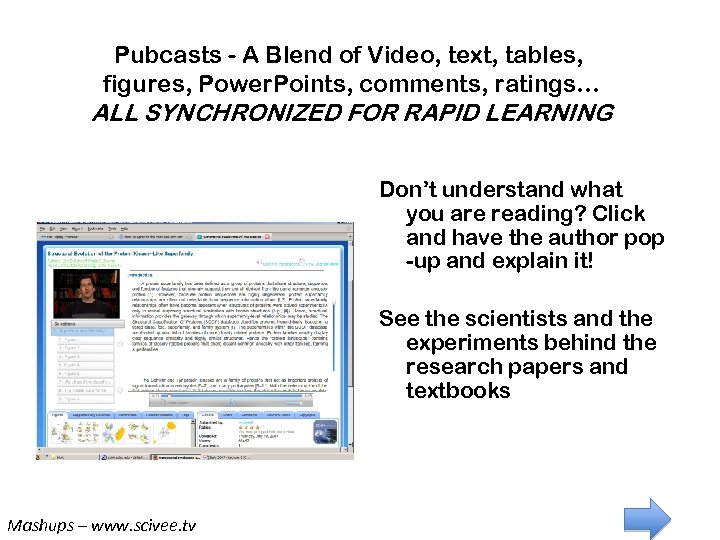 Pubcasts - A Unique Technology Pubcasts - A Blend of Video, text, tables, figures, Power. Points, comments, ratings… ALL SYNCHRONIZED FOR RAPID LEARNING Don’t understand what you are reading? Click and have the author pop -up and explain it! See the scientists and the experiments behind the research papers and textbooks Mashups – www. scivee. tv
Pubcasts - A Unique Technology Pubcasts - A Blend of Video, text, tables, figures, Power. Points, comments, ratings… ALL SYNCHRONIZED FOR RAPID LEARNING Don’t understand what you are reading? Click and have the author pop -up and explain it! See the scientists and the experiments behind the research papers and textbooks Mashups – www. scivee. tv
 Where we Need {Better} Ontologies 2. To Support Tagging of all Aspects of the Scholarly Product
Where we Need {Better} Ontologies 2. To Support Tagging of all Aspects of the Scholarly Product
![Consider Today’s Academic Workflow Reviews Curation Feds Research [Grants] Journal Article Publishers Poster Session Consider Today’s Academic Workflow Reviews Curation Feds Research [Grants] Journal Article Publishers Poster Session](https://present5.com/presentation/2a342ee629e578a29bd4c84b0bffee20/image-34.jpg) Consider Today’s Academic Workflow Reviews Curation Feds Research [Grants] Journal Article Publishers Poster Session Conference Paper Community Service/Data What Should be Done? Societies Blogs
Consider Today’s Academic Workflow Reviews Curation Feds Research [Grants] Journal Article Publishers Poster Session Conference Paper Community Service/Data What Should be Done? Societies Blogs
![Consider Tomorrow’s Academic Workflow Reviews Curation Feds Ideas, Data, Hypotheses Research [Grants] Journal Article Consider Tomorrow’s Academic Workflow Reviews Curation Feds Ideas, Data, Hypotheses Research [Grants] Journal Article](https://present5.com/presentation/2a342ee629e578a29bd4c84b0bffee20/image-35.jpg) Consider Tomorrow’s Academic Workflow Reviews Curation Feds Ideas, Data, Hypotheses Research [Grants] Journal Article Publishers Poster Session Conference Paper Community Service/Data What Should be Done? Societies Blogs
Consider Tomorrow’s Academic Workflow Reviews Curation Feds Ideas, Data, Hypotheses Research [Grants] Journal Article Publishers Poster Session Conference Paper Community Service/Data What Should be Done? Societies Blogs
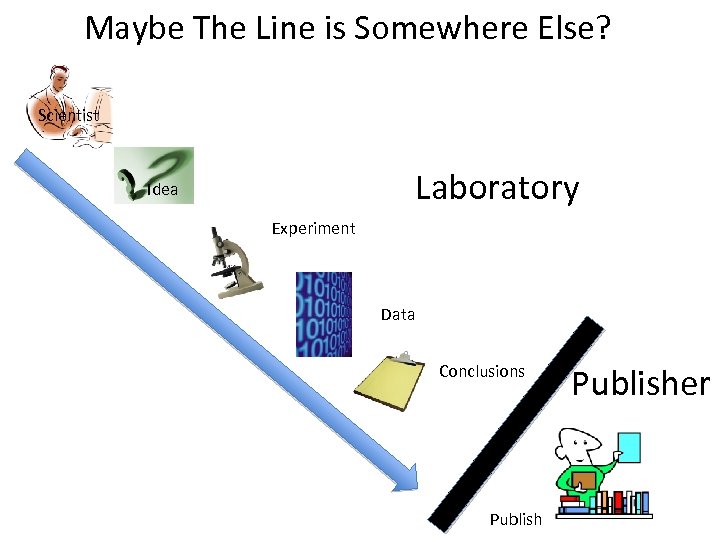 Maybe The Line is Somewhere Else? Scientist Laboratory Idea Experiment Data Conclusions Publisher
Maybe The Line is Somewhere Else? Scientist Laboratory Idea Experiment Data Conclusions Publisher
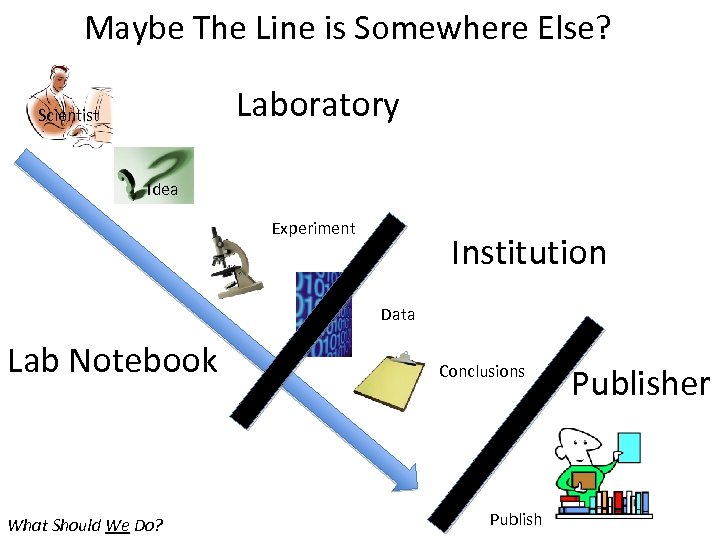 Maybe The Line is Somewhere Else? Laboratory Scientist Idea Experiment Institution Data Lab Notebook What Should We Do? Conclusions Publisher
Maybe The Line is Somewhere Else? Laboratory Scientist Idea Experiment Institution Data Lab Notebook What Should We Do? Conclusions Publisher
 Crowd Sourcing the Electronic Printing Press (aka Workshop: Beyond the PDF) • Proposal to the US National Science Foundation: • Aims: – Define user requirements – Establish a specification document – Open source the development effort – Have a commitment from a publisher to publish a research object using the system – Act as an exemplar for what can be done
Crowd Sourcing the Electronic Printing Press (aka Workshop: Beyond the PDF) • Proposal to the US National Science Foundation: • Aims: – Define user requirements – Establish a specification document – Open source the development effort – Have a commitment from a publisher to publish a research object using the system – Act as an exemplar for what can be done
 Question: What if Everyone Had An Electronic Printing Press? • • • Peer review might change? Bibliometrics might change? Business models will likely change? What happens to the database/literature divide? Societies might do more self publishing? We might have improved the dissemination of science, but will we have improved the comprehension?
Question: What if Everyone Had An Electronic Printing Press? • • • Peer review might change? Bibliometrics might change? Business models will likely change? What happens to the database/literature divide? Societies might do more self publishing? We might have improved the dissemination of science, but will we have improved the comprehension?
 General References • What Do I Want from the Publisher of the Future PLo. S Comp Biol http: //www. sdsc. edu/pb • Fourth Paradigm: Data Intensive Scientific Discovery http: //research. microsoft. com/enus/collabora tion/fourthparadigm/
General References • What Do I Want from the Publisher of the Future PLo. S Comp Biol http: //www. sdsc. edu/pb • Fourth Paradigm: Data Intensive Scientific Discovery http: //research. microsoft. com/enus/collabora tion/fourthparadigm/
 References to Exemplars • Semantic Biochemical Journal - 2010: Using Utopia • Article of the Future, Cell, 2009:
• Prospect, Royal Society of Chemistry, 2009:
• Adventures in Semantic Publishing, Oxford U, 2009: • The Structured Digital Abstract, Seringhaus/Gerstein, 2008
• CWA Nanopublications – 2010
References to Exemplars • Semantic Biochemical Journal - 2010: Using Utopia • Article of the Future, Cell, 2009:
• Prospect, Royal Society of Chemistry, 2009:
• Adventures in Semantic Publishing, Oxford U, 2009: • The Structured Digital Abstract, Seringhaus/Gerstein, 2008
• CWA Nanopublications – 2010
 Acknowledgements • Bio. Lit Team – – – Lynn Fink Parker Williams Marco Martinez Rahul Chandran Greg Quinn • Microsoft Scholarly Communications – – – Pablo Fernicola Lee Dirks Savas Parastitidas Alex Wade Tony Hey http: //www. codeplex. com/ucsdbiolit http//www. pdb. org http: //www. codeplex. com/ucsdbiolit • ww. PDB team • Sci. Vee Team – Apryl Bailey – Tim Beck – – – http: //www. scivee. tv Leo Chalupa Lynn Fink Marc Friedman (CEO) Ken Liu Alex Ramos Willy Suwanto
Acknowledgements • Bio. Lit Team – – – Lynn Fink Parker Williams Marco Martinez Rahul Chandran Greg Quinn • Microsoft Scholarly Communications – – – Pablo Fernicola Lee Dirks Savas Parastitidas Alex Wade Tony Hey http: //www. codeplex. com/ucsdbiolit http//www. pdb. org http: //www. codeplex. com/ucsdbiolit • ww. PDB team • Sci. Vee Team – Apryl Bailey – Tim Beck – – – http: //www. scivee. tv Leo Chalupa Lynn Fink Marc Friedman (CEO) Ken Liu Alex Ramos Willy Suwanto
 pbourne@ucsd. edu Questions?
pbourne@ucsd. edu Questions?


