8ee4c5306f11e67be1d898763e9b6e4b.ppt
- Количество слайдов: 31

The Open Notebook (The Encyclopedia of Life (EOL)) The Annotation and Cataloging of Proteins, Life's Building Blocks for… medicine research education
A Multitude of Data Sites
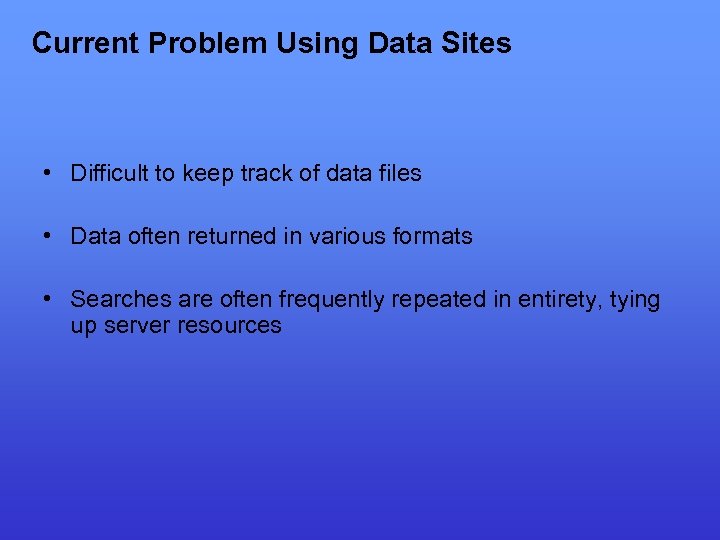
Current Problem Using Data Sites • Difficult to keep track of data files • Data often returned in various formats • Searches are often frequently repeated in entirety, tying up server resources
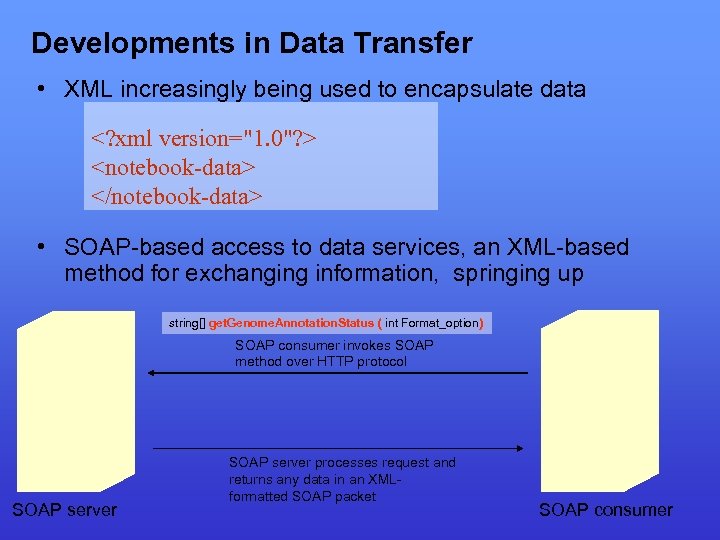
Developments in Data Transfer • XML increasingly being used to encapsulate data <? xml version="1. 0"? > <notebook-data> </notebook-data> • SOAP-based access to data services, an XML-based method for exchanging information, springing up string[] get. Genome. Annotation. Status ( int Format_option) SOAP consumer invokes SOAP method over HTTP protocol SOAP server processes request and returns any data in an XMLformatted SOAP packet SOAP consumer
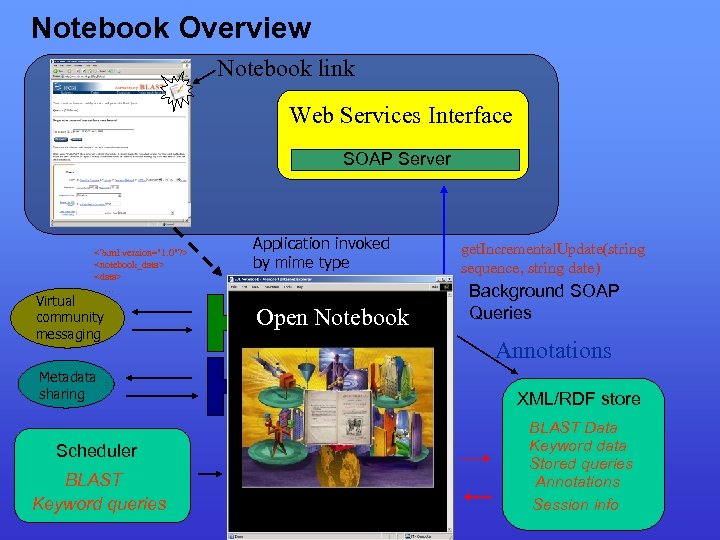
Notebook Overview Notebook link Web Services Interface SOAP Server <? xml version="1. 0"? > <notebook_data> <data> … Virtual community messaging Metadata sharing Scheduler BLAST Keyword queries Application invoked by mime type Open Notebook get. Incremental. Update(string sequence, string date) Background SOAP Queries Annotations XML/RDF store BLAST Data Keyword data Stored queries Annotations Session info

Open Notebook Protocol • Agreed set of protocols for invoking and then feeding with data a client-side application to enable client-side data persistence • Not tied to one programming language
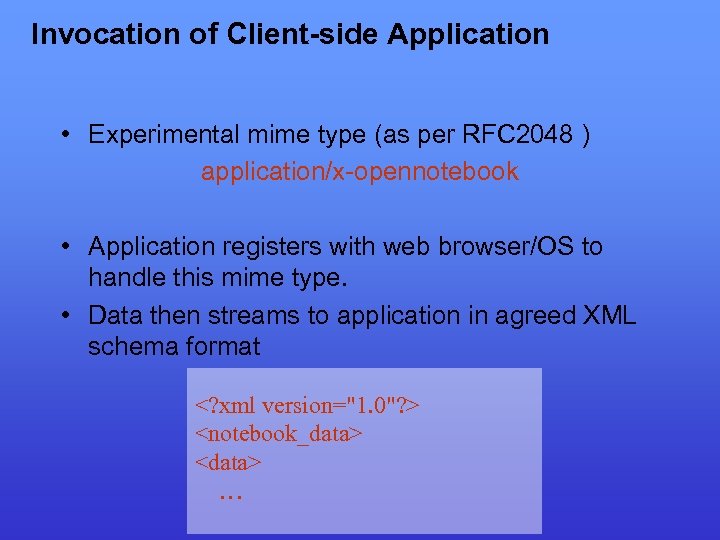
Invocation of Client-side Application • Experimental mime type (as per RFC 2048 ) application/x-opennotebook • Application registers with web browser/OS to handle this mime type. • Data then streams to application in agreed XML schema format <? xml version="1. 0"? > <notebook_data> <data> …

Data would describe required data viewers • Specialized viewers and their current availability specified in XML data download <? xml version="1. 0"? > <notebook_data> <basic-viewer>blast</basic-viewer> <advanced-viewer> <availability>available</availability> <platforms>Java; win 32; macosx</platforms> <download>http: //www. xxx. com/…</download> </advanced-viewer>

Data updates • Indication whether data is updatable <? xml version="1. 0"? > <notebook_data> <updatable>yes</updatable> <SOAP-proxy> http: //www. xxx. org/soapservice< SOAP-proxy> <update-method>get. Genomes(string seq)</update-method> <incrementally-updatable>yes</incrementally-updatable> …

Programming Language-Neutral • Important to just specify protocols and activation scenarios • Enables development of a variety of different and branded versions • Java is envisaged an excellent programming language choice for starting development of an open source version

Encyclopedia of Life • The Encyclopedia of Life (EOL) project is a joint development of the San Diego Supercomputer Center (SDSC) and scientists and biological resources worldwide • EOL involves SDSC staff from HPC (High Performance Computing), DAKS (Distributed Annotation and Knowledge System), Grids, Clusters and Visualization • EOL has three parts: – Putative functional and 3 -D structure assignment through the largest computation ever attempted in biology – Integration of key biological resources – Make this data available to end-user through an intuitive interface • Opportunity to start from ground up
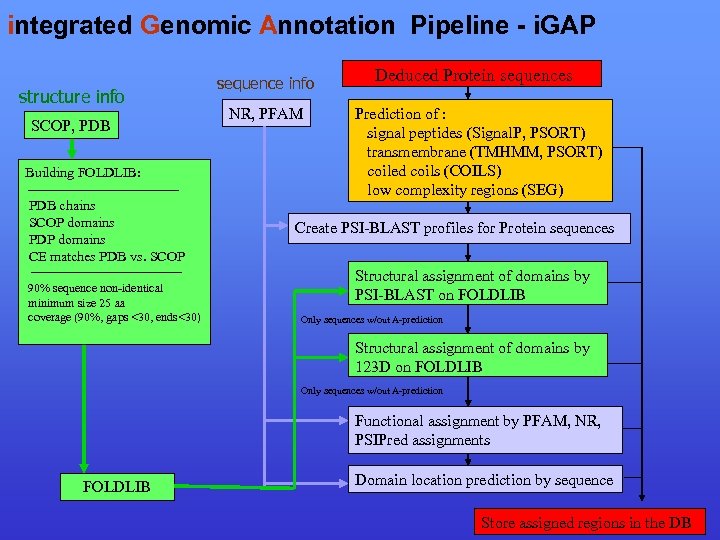
integrated Genomic Annotation Pipeline - i. GAP structure info SCOP, PDB Building FOLDLIB: PDB chains SCOP domains PDP domains CE matches PDB vs. SCOP 90% sequence non-identical minimum size 25 aa coverage (90%, gaps <30, ends<30) sequence info NR, PFAM Deduced Protein sequences Prediction of : signal peptides (Signal. P, PSORT) transmembrane (TMHMM, PSORT) coiled coils (COILS) low complexity regions (SEG) Create PSI-BLAST profiles for Protein sequences Structural assignment of domains by PSI-BLAST on FOLDLIB Only sequences w/out A-prediction Structural assignment of domains by 123 D on FOLDLIB Only sequences w/out A-prediction Functional assignment by PFAM, NR, PSIPred assignments FOLDLIB Domain location prediction by sequence Store assigned regions in the DB
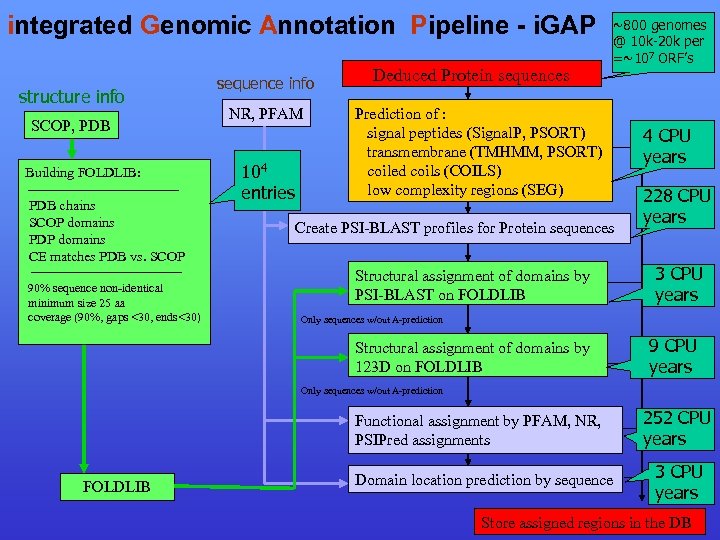
integrated Genomic Annotation Pipeline - i. GAP structure info SCOP, PDB Building FOLDLIB: PDB chains SCOP domains PDP domains CE matches PDB vs. SCOP 90% sequence non-identical minimum size 25 aa coverage (90%, gaps <30, ends<30) sequence info NR, PFAM 104 entries Deduced Protein sequences ~800 genomes @ 10 k-20 k per =~107 ORF’s Prediction of : signal peptides (Signal. P, PSORT) transmembrane (TMHMM, PSORT) coiled coils (COILS) low complexity regions (SEG) Create PSI-BLAST profiles for Protein sequences Structural assignment of domains by PSI-BLAST on FOLDLIB 4 CPU years 228 CPU years 3 CPU years Only sequences w/out A-prediction Structural assignment of domains by 123 D on FOLDLIB 9 CPU years Only sequences w/out A-prediction Functional assignment by PFAM, NR, PSIPred assignments FOLDLIB Domain location prediction by sequence 252 CPU years 3 CPU years Store assigned regions in the DB
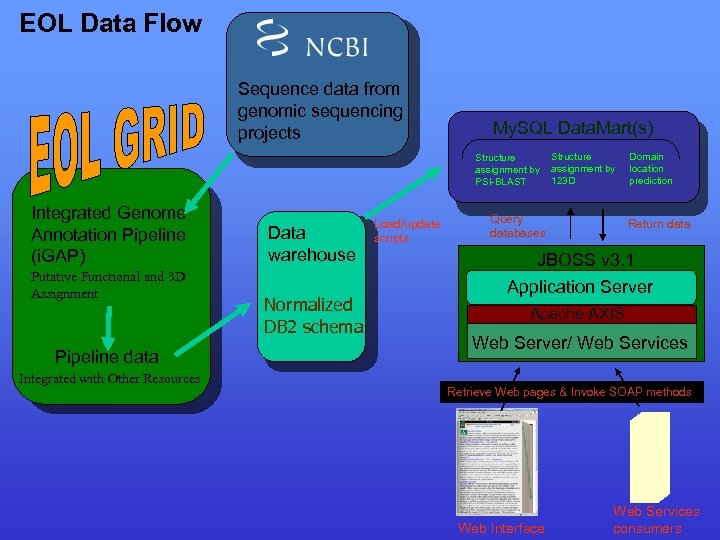
EOL Data Flow Sequence data from genomic sequencing projects My. SQL Data. Mart(s) Structure assignment by PSI-BLAST Integrated Genome Annotation Pipeline (i. GAP) Putative Functional and 3 D Assignment Pipeline data Integrated with Other Resources Data warehouse Normalized DB 2 schema Load/update scripts Structure assignment by 123 D Query databases Domain location prediction Return data JBOSS v 3. 1 Application Server Apache AXIS Web Server/ Web Services Retrieve Web pages & Invoke SOAP methods Web Interface Web Services consumers
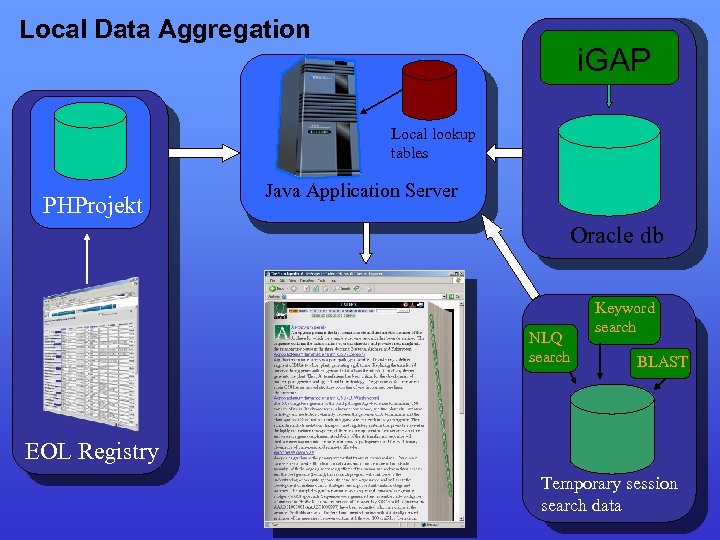
Local Data Aggregation i. GAP Local lookup tables PHProjekt Java Application Server Oracle db NLQ search Keyword search BLAST EOL Registry Temporary session search data
EOL Front End: Web Interface

Interactive Data Rendering • Need for interactive client side graphical data rendering • Flash used in EOL prototype but… – development time high – thin client capabilities limited by player parsing capabilities • Scalable Vector Graphics (SVG) – – Described by an XML-based text file graphic description can be created server-side standards based Interactivity provided by embedded ECMA scripting • Negatives: – Little native support in web browsers – Must use proprietary plugin (Adobe) in practice
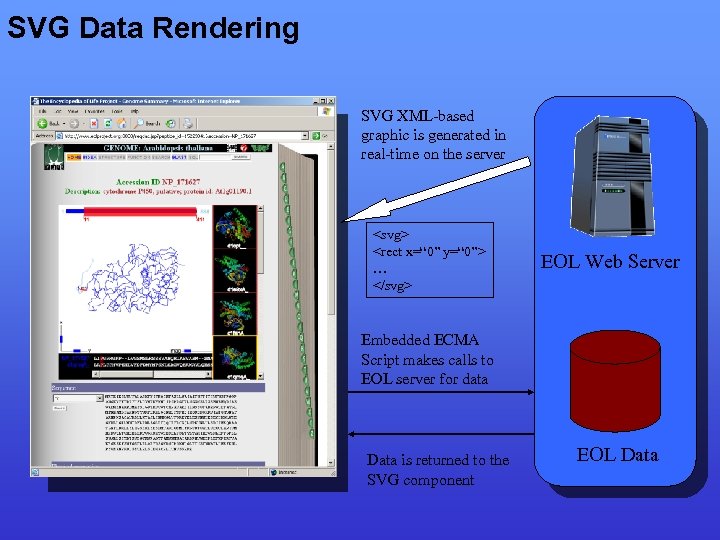
SVG Data Rendering SVG XML-based graphic is generated in real-time on the server <svg> <rect x=“ 0” y=“ 0”> … </svg> EOL Web Server Embedded ECMA Script makes calls to EOL server for data Data is returned to the SVG component EOL Data
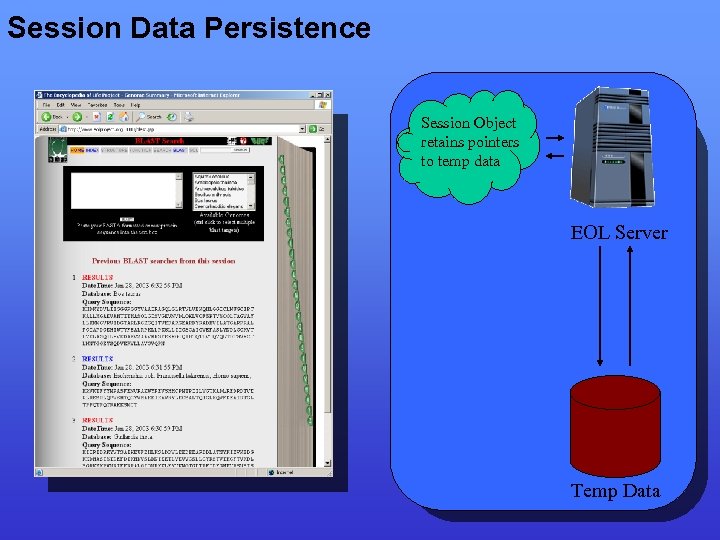
Session Data Persistence Session Object retains pointers to temp data EOL Server Temp Data
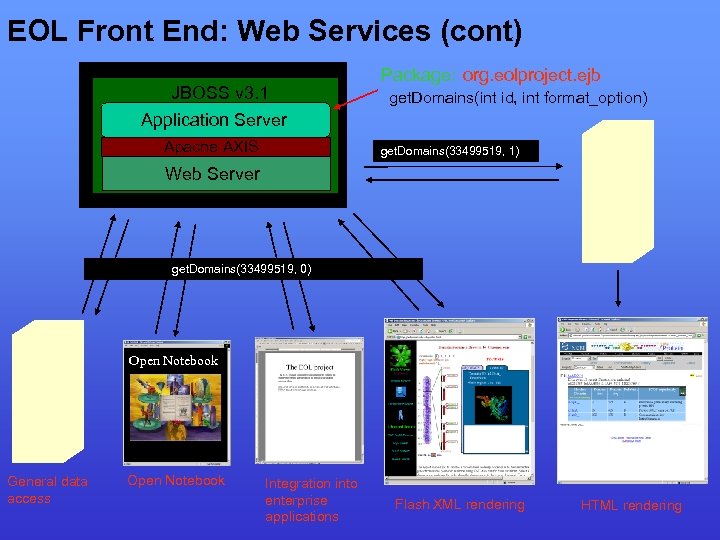
EOL Front End: Web Services (cont) JBOSS v 3. 1 Application Server Apache AXIS Package: org. eolproject. ejb get. Domains(int id, int format_option) get. Domains(33499519, 1) Web Server get. Domains(33499519, 0) Open Notebook General data access Open Notebook Integration into enterprise applications Flash XML rendering HTML rendering

Open Notebook Software Wish List • • • Multi-Platform application Easy installation and update Local search functionality Data annotation Built-in basic data viewers for popular data, i. e. BLAST, sequence alignments, basic molecular rendering Automated download of specialized data viewers Automatic data updates via background use of web services User notification of new data Point-and-click interface to support new breed of PDA’s and Tablets Peer-to-peer querying of annotation data

Easy Installation and Update • Idiot-proof installation • Java Network Launch Protocol (JNLP) good contender, i. e. Web. Start • JNLP has ability to provide application updates

Local search functionality • Whatever kind of database is used, it needs to be able to support some kind of search functionality • For the open notebook project we would seek an open source XML-based database, look to xml: db API for a means to interact with a native XML database • EXIST is one example of an open source, native XML database

Data annotation & Peer-to-peer querying of annotation data • Personal annotations on local data a useful and relatively easy feature to implement • Peer-to-peer access contentious and needs to be well controlled • Potentially could create a real community of online scientists • Effectively a scientific “Napster”
Built-in Basic Data Viewers • Need to have minimum built-in capability – – – Text viewer SVG Graphics viewer NCBI DTD-based BLAST browser Multiple sequence alignment viewer Molecule renderer

Automatic data updates via SOAP calls • Server-side must be set up for providing SOAP method calls • Potential to drastically reduce server load by performing incremental search get. Blast. Data( string sequence, string last-queried )

Point-and-click interface • Intuitive interface • Constructed with an eye on developments in personal computing e. g. PDA’s and Tablet computers

What Next…? • Upload a seed Java-based project onto the Bioinformatics. org site together with an RFC • Discuss online the merits of the project

Summary • A genuine need for a means to: – – Collate data Automatic updates of data Enable shared data annotations Specialized data processing • Java provides a compelling platform to develop an open version of this client-side application

EOL Team Dave Archbell Kim Baldridge Chaitanya Baru Fran Berman Philip Bourne Robert Byrnes Henri Casanova Eliot Clingman Neil Cotofana Cassie Ferguson Tony Fountain Jerry Greenberg Michael Gribskov Dana Jermanis Wilfred Li Jennifer Matthews Mark Miller Julie Mitchell Coleman Mosley Greg Quinn Vicente Reyes Jerry Rowley Peter Shin Ilya Shindyalov Chris Smith David Stoner Stella Veretnik

Further information: http: //www. eolproject. info http: //www. bioinformatics. org/opennotebook
8ee4c5306f11e67be1d898763e9b6e4b.ppt