579b5ff610679cbb3c3db9fb5ad39662.ppt
- Количество слайдов: 32
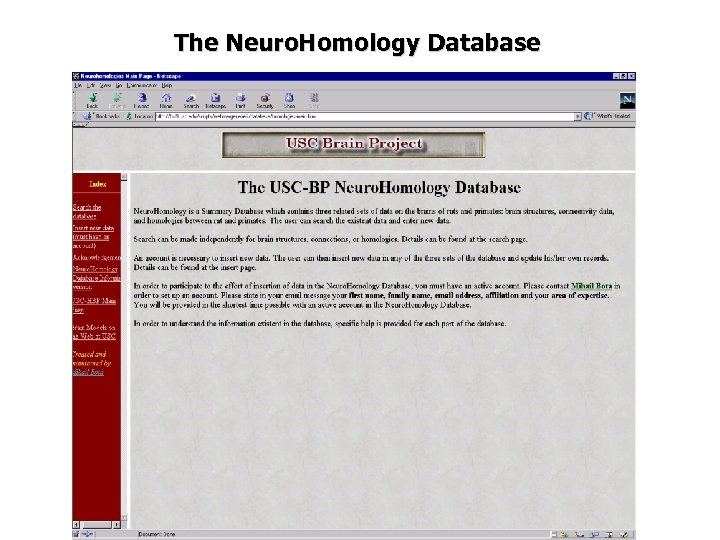
The Neuro. Homology Database
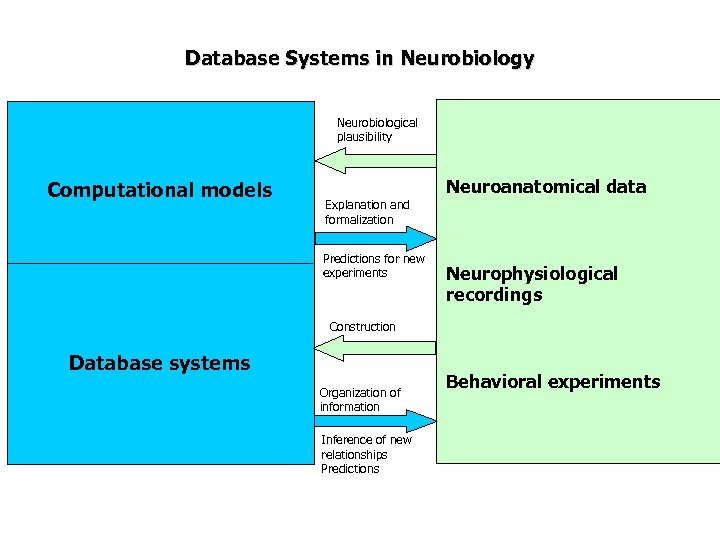
Database Systems in Neurobiology Neurobiological plausibility Computational models Neuroanatomical data Explanation and formalization Predictions for new experiments Neurophysiological recordings Construction Database systems Organization of information Inference of new relationships Predictions Behavioral experiments
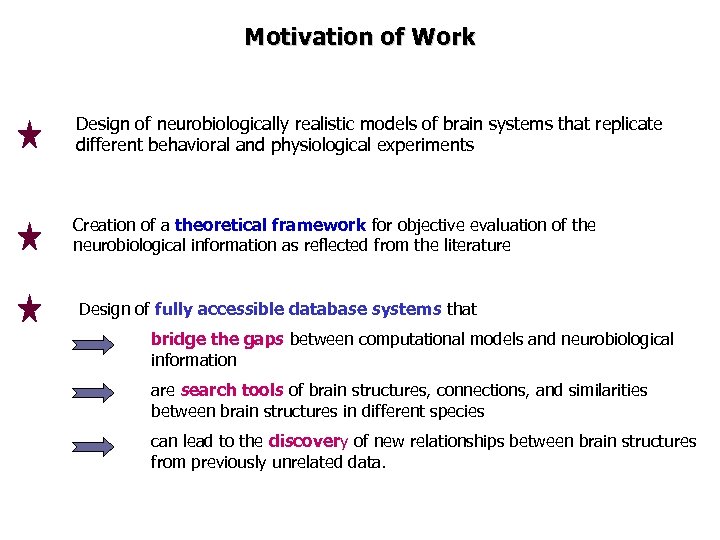
Motivation of Work Design of neurobiologically realistic models of brain systems that replicate different behavioral and physiological experiments Creation of a theoretical framework for objective evaluation of the neurobiological information as reflected from the literature Design of fully accessible database systems that bridge the gaps between computational models and neurobiological information are search tools of brain structures, connections, and similarities between brain structures in different species can lead to the discovery of new relationships between brain structures from previously unrelated data.
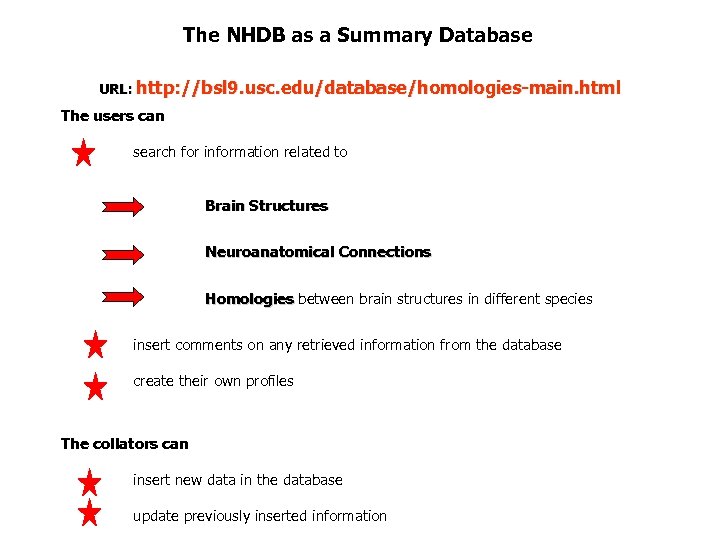
The NHDB as a Summary Database URL: http: //bsl 9. usc. edu/database/homologies-main. html The users can search for information related to Brain Structures Neuroanatomical Connections Homologies between brain structures in different species insert comments on any retrieved information from the database create their own profiles The collators can insert new data in the database update previously inserted information
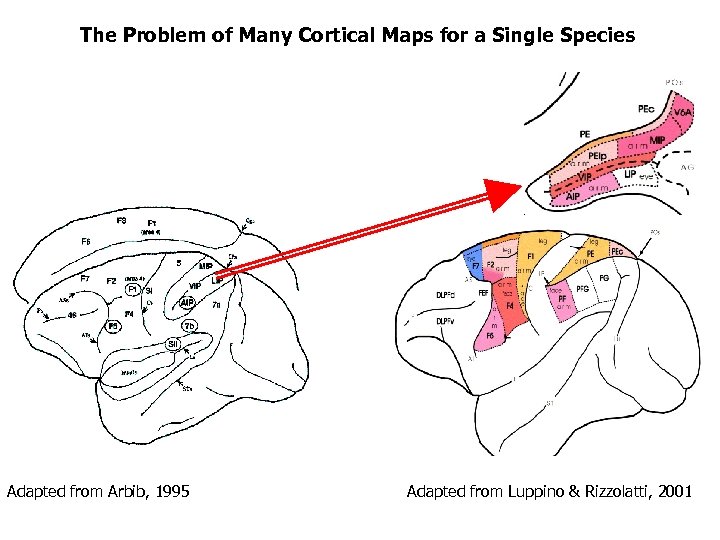
The Problem of Many Cortical Maps for a Single Species Adapted from Arbib, 1995 Adapted from Luppino & Rizzolatti, 2001
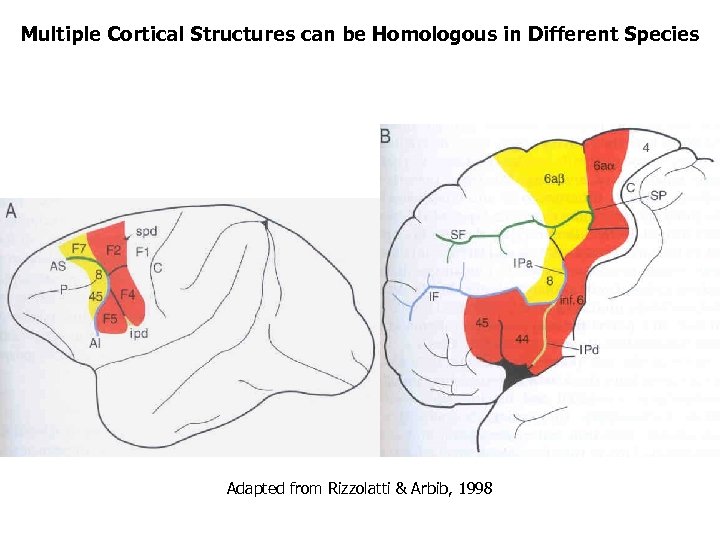
Multiple Cortical Structures can be Homologous in Different Species Adapted from Rizzolatti & Arbib, 1998
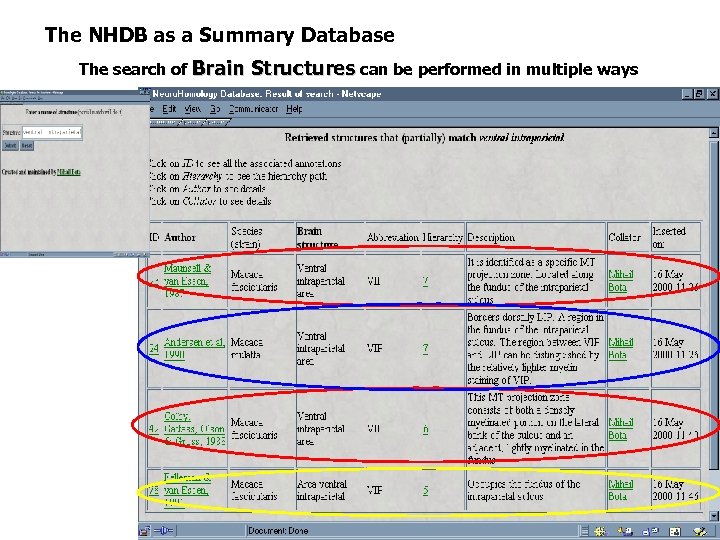
The NHDB as a Summary Database The search of Brain Structures can be performed in multiple ways
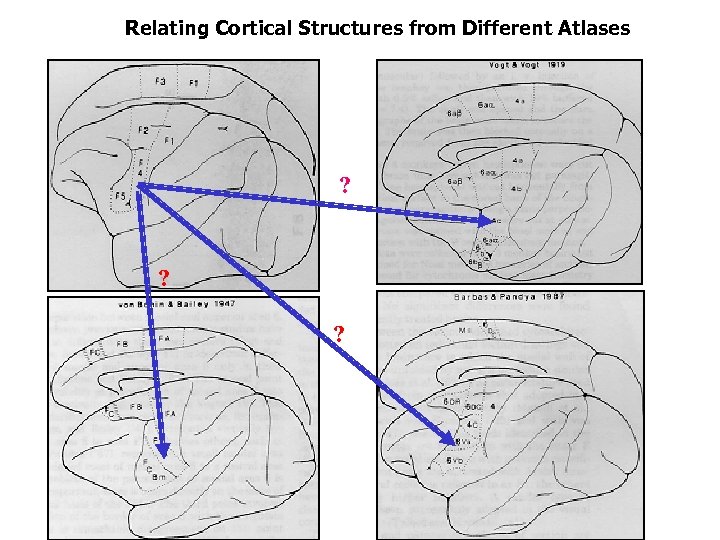
Relating Cortical Structures from Different Atlases ? ? ?
Relating Cortical Structures from Different Atlases The translation inference engine exploits a qualitative spatial inference algorithm developed in the GIS paradigm evaluates the neuroanatomical connections of cortical structures in different parcellation schemes can be used for reconstruction of the patterns of connectivity of cortical structures in a given brain atlas, from the connectivity information obtained in different atlases
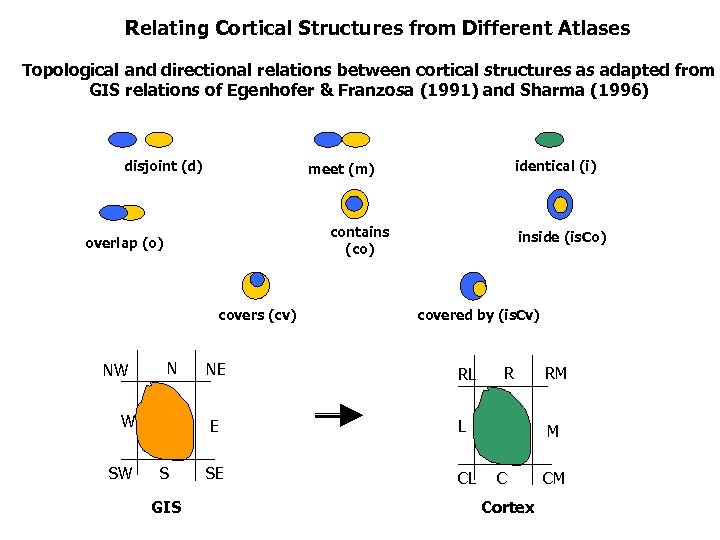
Relating Cortical Structures from Different Atlases Topological and directional relations between cortical structures as adapted from GIS relations of Egenhofer & Franzosa (1991) and Sharma (1996) disjoint (d) contains (co) overlap (o) covers (cv) NW N inside (is. Co) covered by (is. Cv) S GIS NE RL E W SW identical (i) meet (m) L SE CL R RM M C Cortex CM
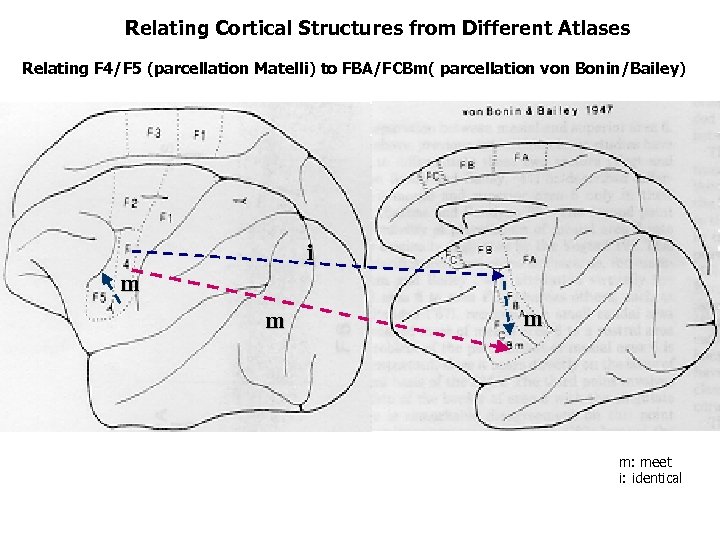
Relating Cortical Structures from Different Atlases Relating F 4/F 5 (parcellation Matelli) to FBA/FCBm( parcellation von Bonin/Bailey) i m m: meet i: identical
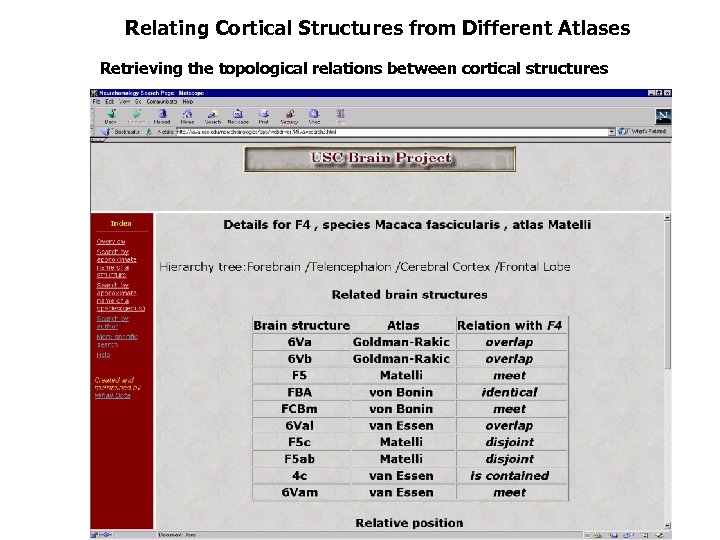
Relating Cortical Structures from Different Atlases Retrieving the topological relations between cortical structures
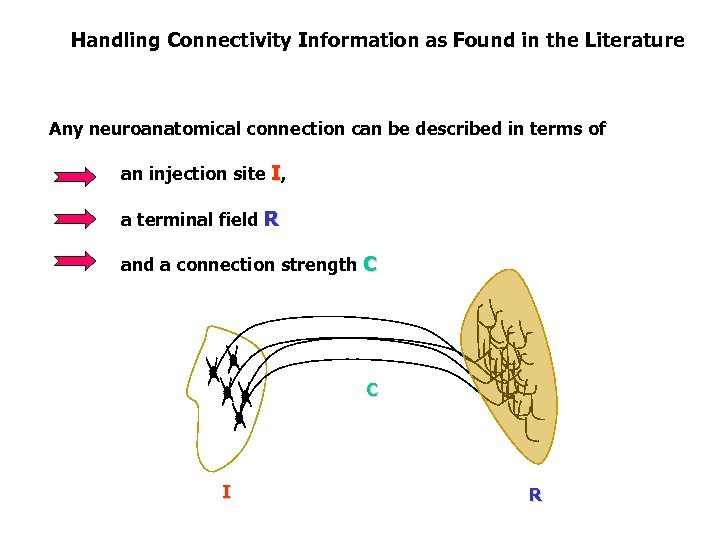
Handling Connectivity Information as Found in the Literature Any neuroanatomical connection can be described in terms of an injection site I, a terminal field R and a connection strength C C I R

The Search of Connections can be Performed in Multiple Ways
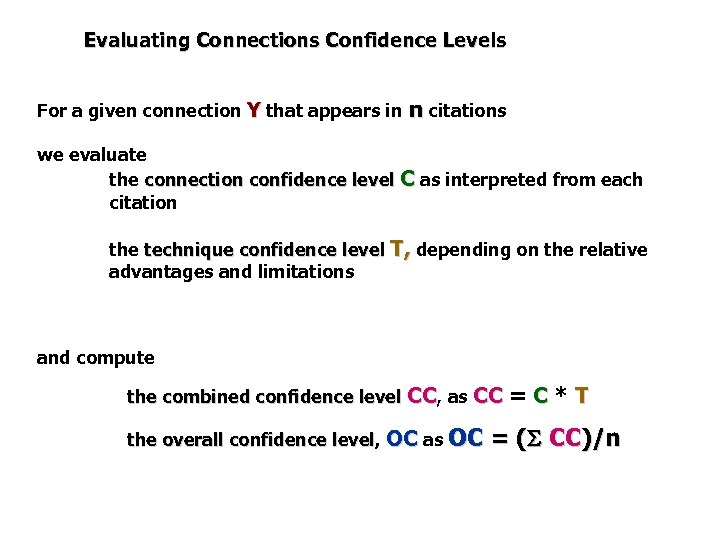
Evaluating Connections Confidence Levels For a given connection Y that appears in n citations we evaluate the connection confidence level C as interpreted from each citation the technique confidence level T, depending on the relative advantages and limitations and compute the combined confidence level CC, as CC the overall confidence level, OC as OC level =C*T = ( CC)/n
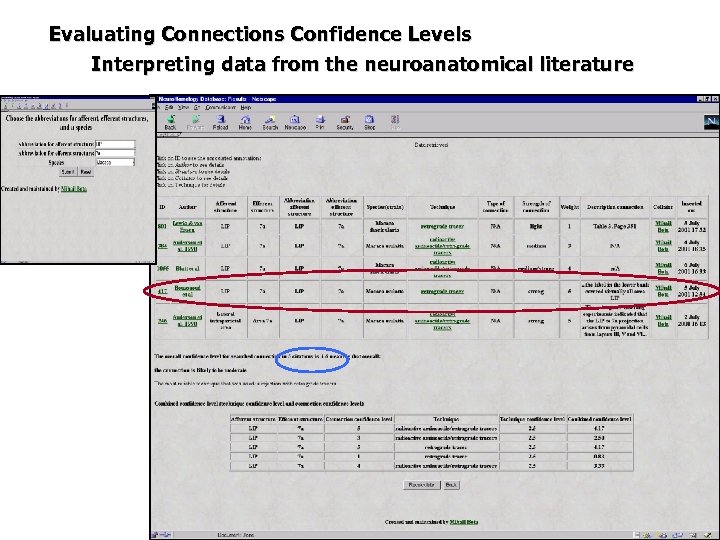
Evaluating Connections Confidence Levels Interpreting data from the neuroanatomical literature
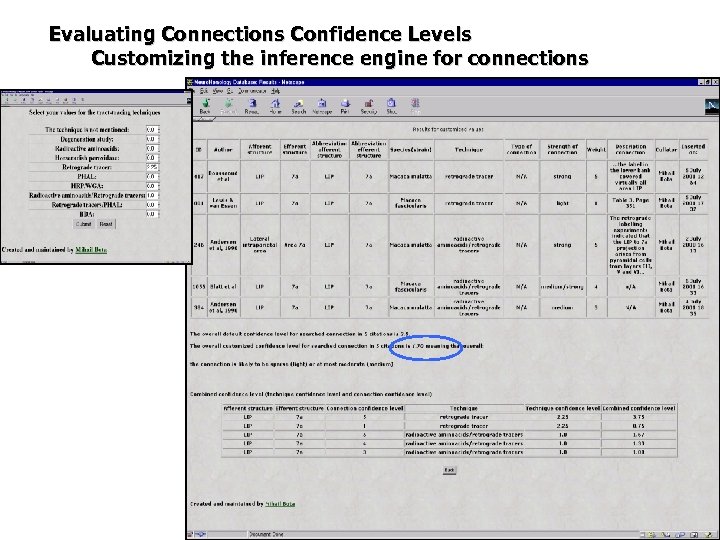
Evaluating Connections Confidence Levels Customizing the inference engine for connections
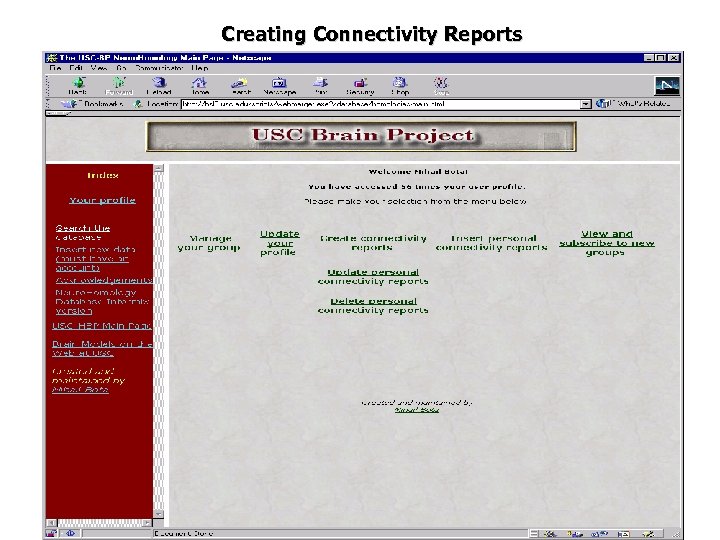
Creating Connectivity Reports
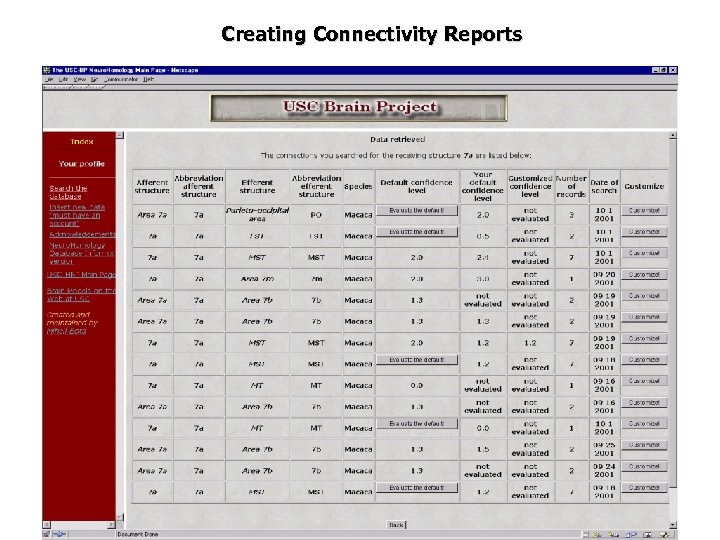
Creating Connectivity Reports
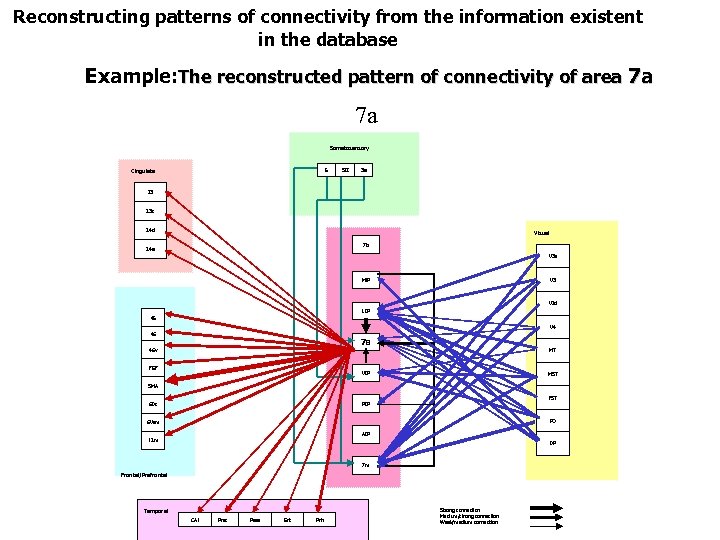
Reconstructing patterns of connectivity from the information existent in the database Example: The reconstructed pattern of connectivity of area 7 a 7 a Somatosensory 5 Cingulate SII 3 a 23 23 c 24 d Visual 7 b 24 a V 3 MIP V 2 d LIP 45 V 4 46 7 a 46 v MT FEF VIP MST SMA FST PIP 6 Ds PO 6 Vam AIP 12 m DP 7 m Frontal/Prefrontal Temporal CA 1 Pres Para Ent Prh Strong connection Medium/strong connection Weak/medium connection
The Concept of Homology The structuralist approach: there is a common structural plan across vertebrates (“Bauplan”) The phylogenetic approach: characters have to be followed across species. Hierarchies of structures within the nervous system: cellular aggregates, composed of major brain regions, brain nuclei and nuclear subdivisions cell types, including major types and subtypes molecules grouped into families and superfamilies
Evaluating Homologies between Brain Structures the degree of homology of a pair of brain structures from different species depends on how close are those nuclei which are related with the compared structures. the evaluation of the degree of homology depends also on the reliability of information inserted in the database users can perform customized evaluations of the degree of homology users can comparatively evaluate the degrees of homology of a brain structure from a given species with a number of different structures from another species

The Concept of Degree of Homology We propose the concept of degree of homology DG as an overall measure for how close two brain structures from different species are. The criteria we use for evaluating the degree of homology : relative position afferent connections efferent connections chemoarchitecture cell morphology gross appearance myeloarchitecture functionality For each of the considered criteria, an index of similarity IS is associated
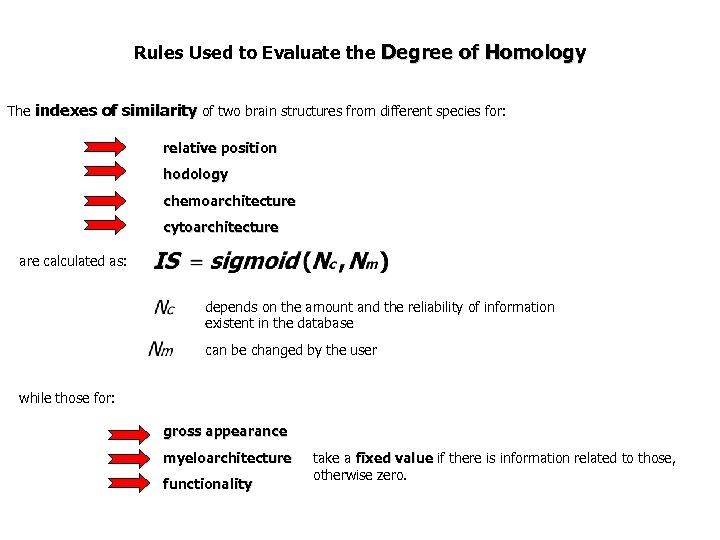
Rules Used to Evaluate the Degree of Homology The indexes of similarity of two brain structures from different species for: relative position hodology chemoarchitecture cytoarchitecture are calculated as: depends on the amount and the reliability of information existent in the database can be changed by the user while those for: gross appearance myeloarchitecture functionality take a fixed value if there is information related to those, otherwise zero.
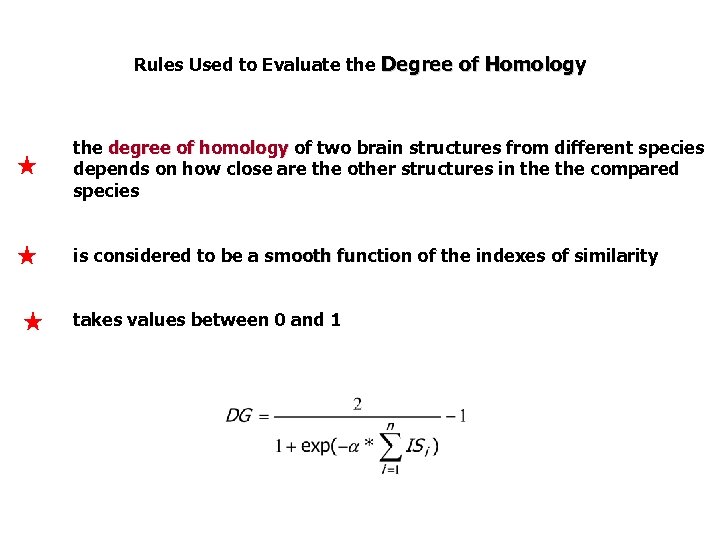
Rules Used to Evaluate the Degree of Homology the degree of homology of two brain structures from different species depends on how close are the other structures in the compared species is considered to be a smooth function of the indexes of similarity takes values between 0 and 1
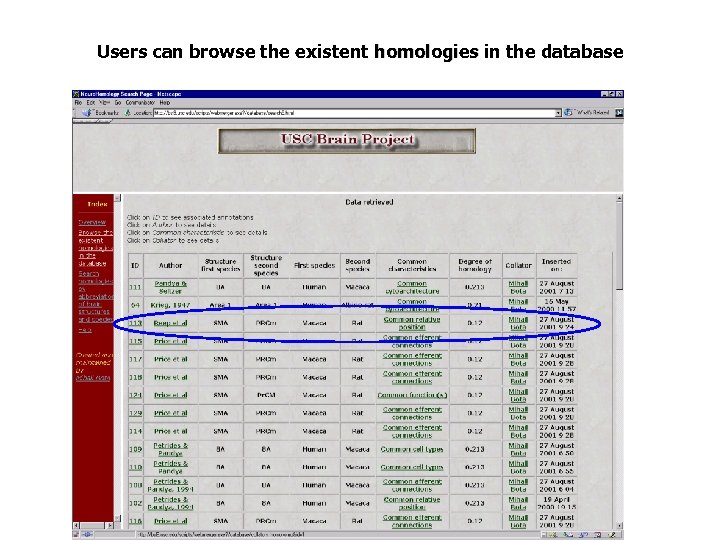
Users can browse the existent homologies in the database
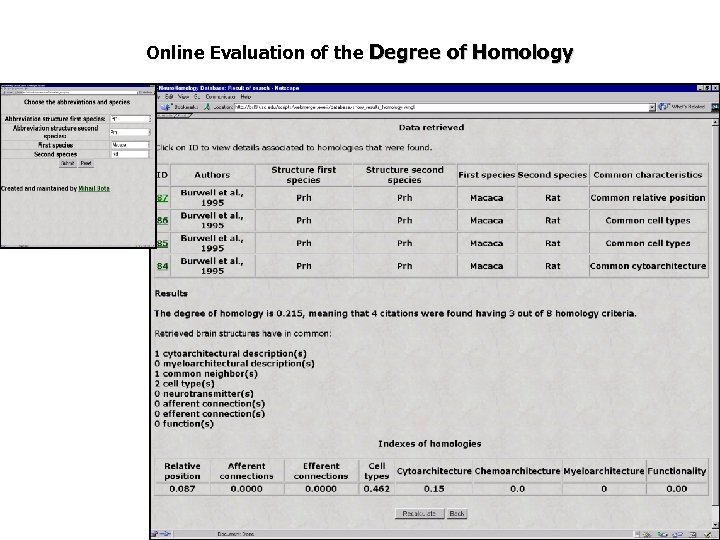
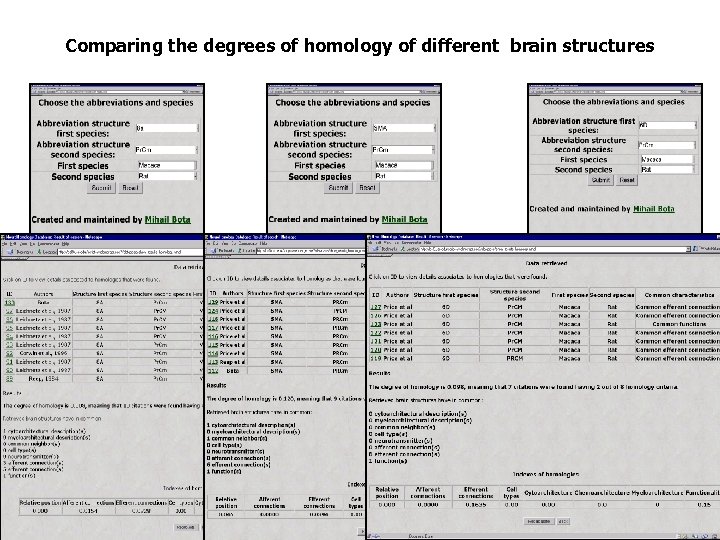
Online Evaluation of the Degree of Homology
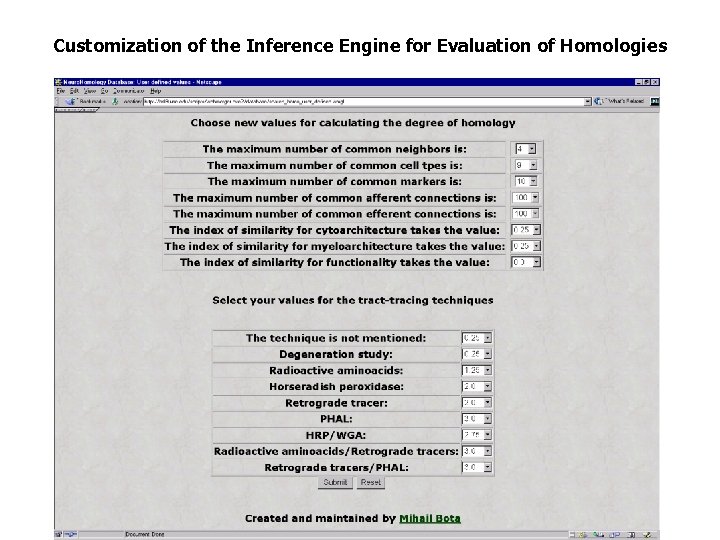
Customization of the Inference Engine for Evaluation of Homologies
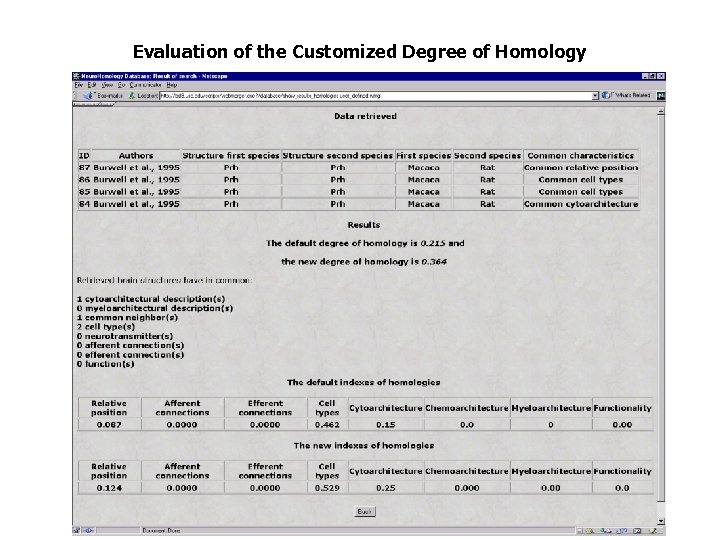
Evaluation of the Customized Degree of Homology
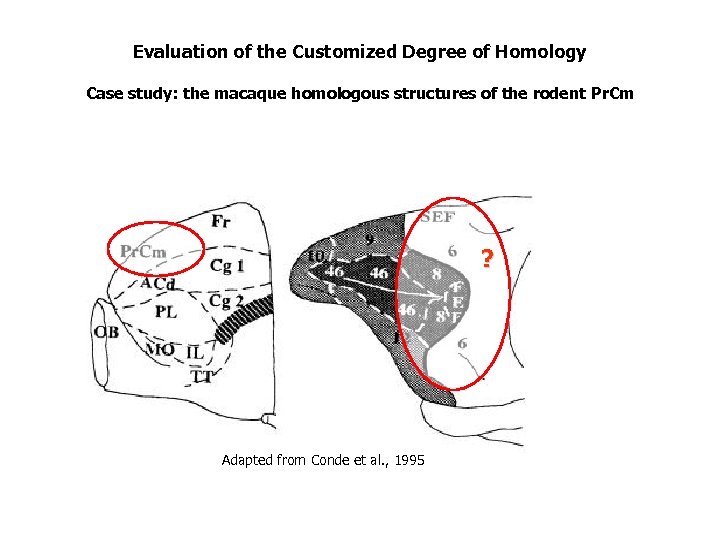
Evaluation of the Customized Degree of Homology Case study: the macaque homologous structures of the rodent Pr. Cm ? Adapted from Conde et al. , 1995
Comparing the degrees of homology of different brain structures

Conclusions and Future Work We designed a set of database systems that can be accessed online both in terms of searching for information and to insert new data The structures of the designed database systems allow the members of the neuroscientific community to use them both as summary databases for neurobiological information and also as expert systems for inference of new relationships from previously unrelated data The expert systems can lead to predictions of new functional parcellations of cortical structures and may be used for construction of hierarchies of homologous structures. Refinement of the existent expert systems Inclusion of more homology criteria Automated creation of the connectivity matrices of brain structures of interest.
579b5ff610679cbb3c3db9fb5ad39662.ppt