9b8aebba0916402af1e7f79f6be3f710.ppt
- Количество слайдов: 54

The i. Plant Collaborative Cyberinfrastructure aka Development of Public Cyberinfrastructure to Support Plant Science Presented by Dan Stanzione Co-PI and Cyberinfrastructure Lead, i. Plant Collaborative Deputy Director, Texas Advanced Computing Center

Today’s Schedule • Presentations – Me: Overview and architecture – Matt: CI for Genotype to Phenotype – Sheldon: CI for Tree of Life – Uwe: CI for Education the next generation of biologists. • A quick break • Live interactive demos of DNA Subway, Discovery Environment, and My. Plant site.

What is i. Plant? • i. Plant’s mission is to build the CI to support plant biology’s Grand Challenge solutions • Grand Challenges were not defined in advance, but identified through engagement with the community • A virtual organization with Grand Challenge teams relying on national cyberinfrastructure • Long term focus on sustainable food supply, climate change, biofuels, ecological stability, etc • Hundreds of participants globally… Working group members at >50 US institutions, USDA, DOE, etc.

Brief History • Formally approved by National Science Board – 12/2007 • Funding by NSF – February 1 st, 2008 • i. Plant Kickoff Conference at CSHL – April 2008 o ~200 participants Ø Grand Challenge Workshops – Sept-Dec 2008 Ø CI workshop – Jan 2009 Ø Grand Challenge White Paper Review – March 2009 Ø Project Recommendations – March 2009 Ø Project Kickoffs – May 2009 & August 2009 Ø First Release of Discovery Environments – April 2010

Grand Challenge & CI Workshops • Mechanistic Basis of Plant Adaptation (9 -30 -08) • Impact of Climate Change on Plant Productivity: Prediction of Phenotype from Genotype (9 -30 -08) • Developing common models for molecular mechanisms, crop physiology, and ecology (11 -7 -08) • Assembling the Tree of Life to Enable the Plant Sciences (11 -19 -08) • Computational Morphodynamics of Plants (12 -15 -08) • Botanical Information & Ecology Network • CI Workshop

GC Projects Recommended by the i. Plant Board of Directors March 2009 Initial Projects: Plant Tree of Life – i. PTo. L – May ‘ 09 +Taxonomic Intelligence + APWeb 2 + Social Networking Website Genotype to Phenotype – i. PG 2 P – Aug ‘ 09 + Image Analysis Platform
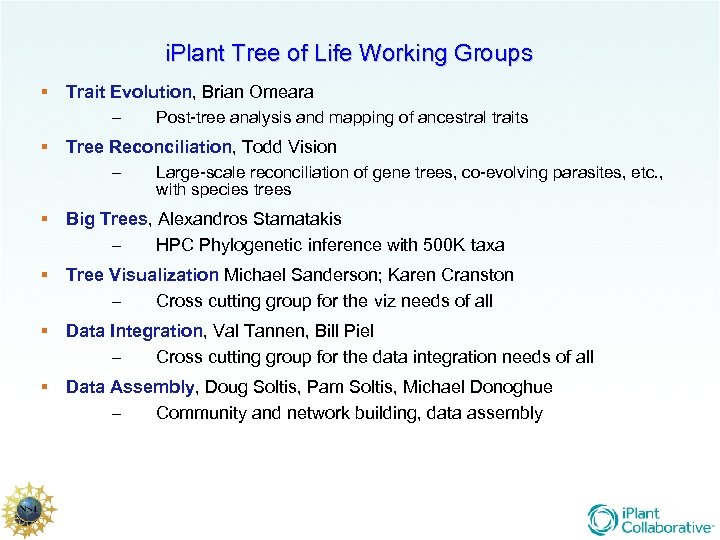
i. Plant Tree of Life Working Groups Trait Evolution, Brian Omeara – Post-tree analysis and mapping of ancestral traits Tree Reconciliation, Todd Vision – Large-scale reconciliation of gene trees, co-evolving parasites, etc. , with species trees Big Trees, Alexandros Stamatakis – HPC Phylogenetic inference with 500 K taxa Tree Visualization Michael Sanderson; Karen Cranston – Cross cutting group for the viz needs of all Data Integration, Val Tannen, Bill Piel – Cross cutting group for the data integration needs of all Data Assembly, Doug Soltis, Pam Soltis, Michael Donoghue – Community and network building, data assembly
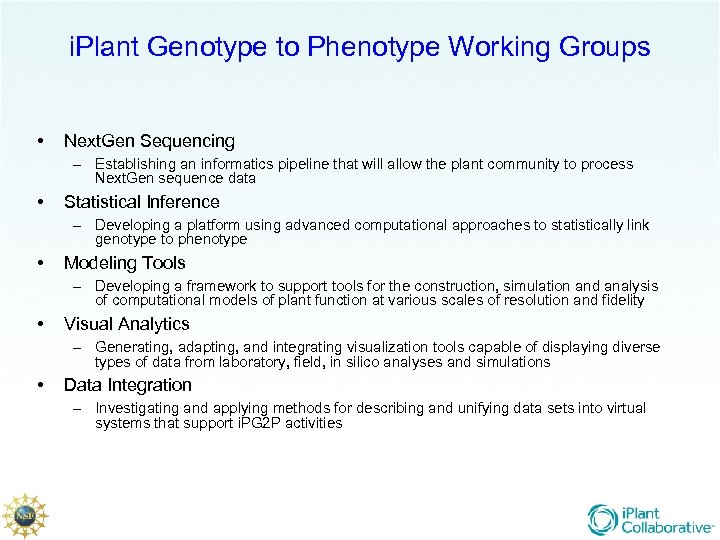
i. Plant Genotype to Phenotype Working Groups • Next. Gen Sequencing – Establishing an informatics pipeline that will allow the plant community to process Next. Gen sequence data • Statistical Inference – Developing a platform using advanced computational approaches to statistically link genotype to phenotype • Modeling Tools – Developing a framework to support tools for the construction, simulation and analysis of computational models of plant function at various scales of resolution and fidelity • Visual Analytics – Generating, adapting, and integrating visualization tools capable of displaying diverse types of data from laboratory, field, in silico analyses and simulations • Data Integration – Investigating and applying methods for describing and unifying data sets into virtual systems that support i. PG 2 P activities
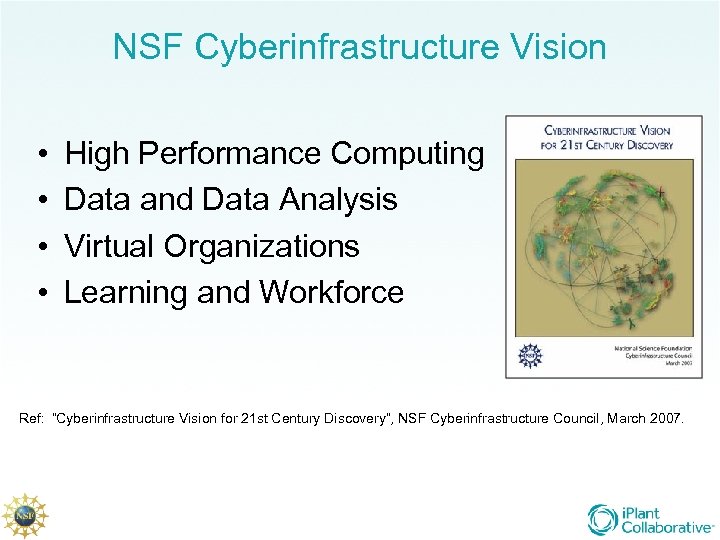
NSF Cyberinfrastructure Vision • • High Performance Computing Data and Data Analysis Virtual Organizations Learning and Workforce Ref: “Cyberinfrastructure Vision for 21 st Century Discovery”, NSF Cyberinfrastructure Council, March 2007.
What is Cyberinfrastructure? (Originally about Tera. Grid) WWW. TERAGRID. ORG It was six men of Indostan, To learning much inclined, Who went to see the elephant, (Though all of them were blind), That each by observation Might satisfy his mind. It’s a Grid! It’s a Network! They are HPC Centers! It’s a Common Software Environ! And More!: - Viz - Facilities - Data collections It’s Apps and Support! It’s Storage! …
Cyberinfrastructure versus Bioinformatics • Leveraging production compute and storage infrastructure; hundreds of millions in NSF investment… these aren’t machines in our lab. • Focus on a *platform* not tools – Methods for leveraging physical resources – Methods for integrating tools – Methods for integrating data • Emphasis on a sustainable, species independent platform.
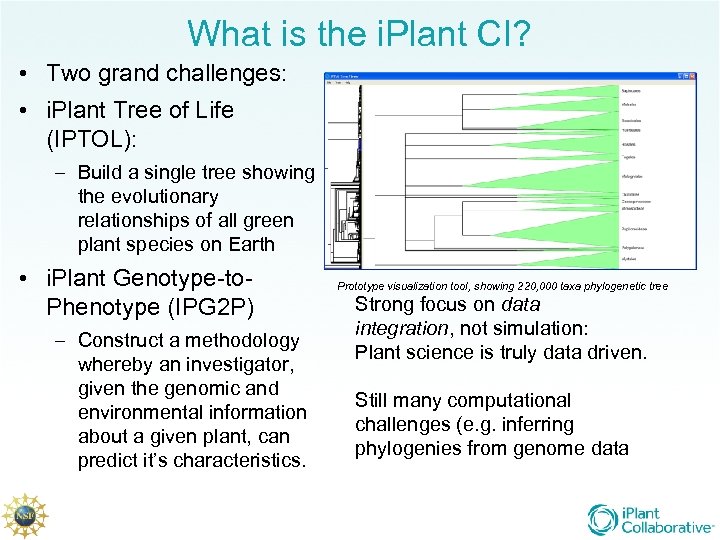
What is the i. Plant CI? • Two grand challenges: • i. Plant Tree of Life (IPTOL): – Build a single tree showing the evolutionary relationships of all green plant species on Earth • i. Plant Genotype-to. Phenotype (IPG 2 P) – Construct a methodology whereby an investigator, given the genomic and environmental information about a given plant, can predict it’s characteristics. Prototype visualization tool, showing 220, 000 taxa phylogenetic tree Strong focus on data integration, not simulation: Plant science is truly data driven. Still many computational challenges (e. g. inferring phylogenies from genome data

Open Source Philosophy, Commercial Quality Process • i. Plant is open in every sense of the word: – Open access to source – Open API to build a community of contributors – Open standards adopted wherever possible – Open access to data (where users so choose). • i. Plant code design, implementation, and quality control will be based in best industrial practice
CI Timelines • Per NSF mandate: No development before conclusion of GC selection in March, 2009 • GC projects kicked off requirements gathering phase in May and July 2009, respectively. • Software engineering practices established, staffing expansion in summer of 2009 • Architecture design first production and prototype coding began in September 2009 • Initial prototype rollouts began Jan. 2010 • First product betas began March 2010 • New releases of DE quarterly, with periodic releases of other products.

IPTOL CI – At a Very High Level • Goal: Build and use very large trees, perhaps all green plant species • Needs: – Most of the data isn’t collected. A lot of what is collected isn’t organized. – Lots of analysis tools exist (probably plenty of them) – but they don’t work together, and use many different data formats. – The tree builder tools take too long to run. – The visualization tools don’t scale to the tree sizes needed.
IPTOL CI – High Level • Addressing these needs through CI – My. Plant – the social networking site for phylogenetic data collection (organized by clade) – Provide a common repository for data without an NCBI home (e. g. 1 k. P) – Discovery Environment: Build a common interface, data format, and API to unite tools. – Enhance tree builder tools (RAx. ML, NINJA, Sate’) with parallelization and checkpointing – Build a remote visualization tool capable of running where we can guarantee RAM resources

Support of Existing Tools • The IPTOL working groups have determined a number of tools that needed to be enhanced to meet initial scientific goals: – NINJA (Neighbor Joining) – RAXML (Maximum Likeklihood) – Both pose significant scalability challenges i. Plant staff are helping the developers tackle.
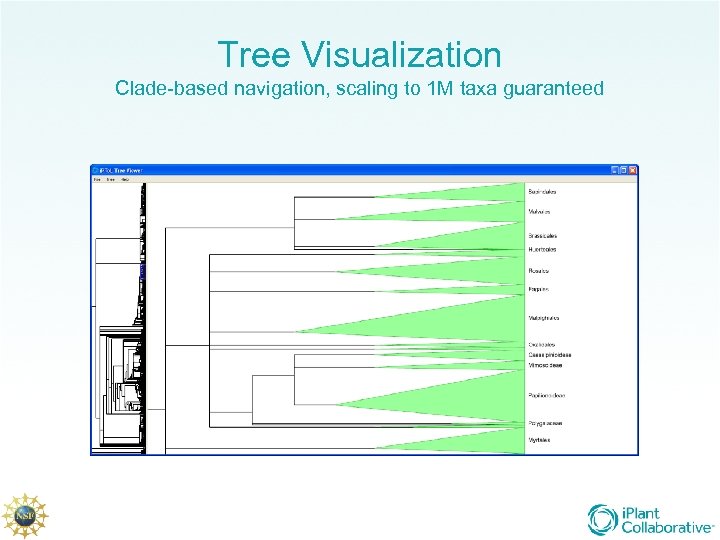
Tree Visualization Clade-based navigation, scaling to 1 M taxa guaranteed
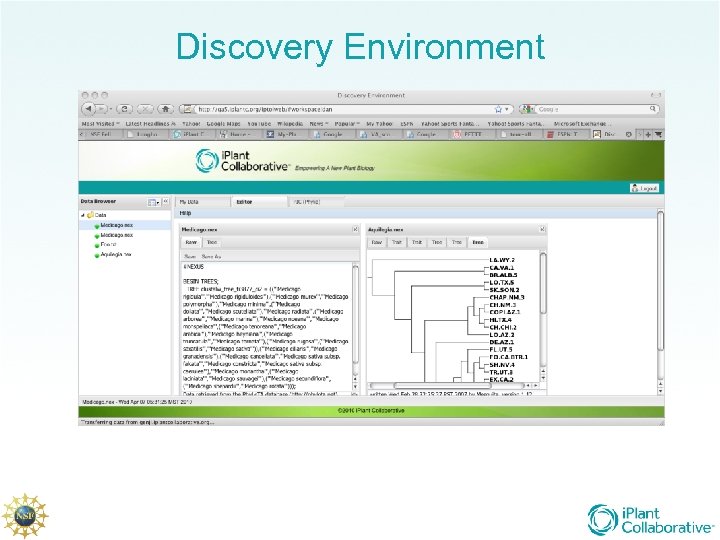
Discovery Environment

First DE • Support of only one workflow, independent contrasts, but: – Remote execution of compute tasks on Tera. Grid resources seamlessly – Incorporation of existing informatics tools behind i. Plant interface – Parsing of multiple data formats into i. Plant format – Seamless integration of online data resources – Role based access and basic provenance support • Mostly foundation work…
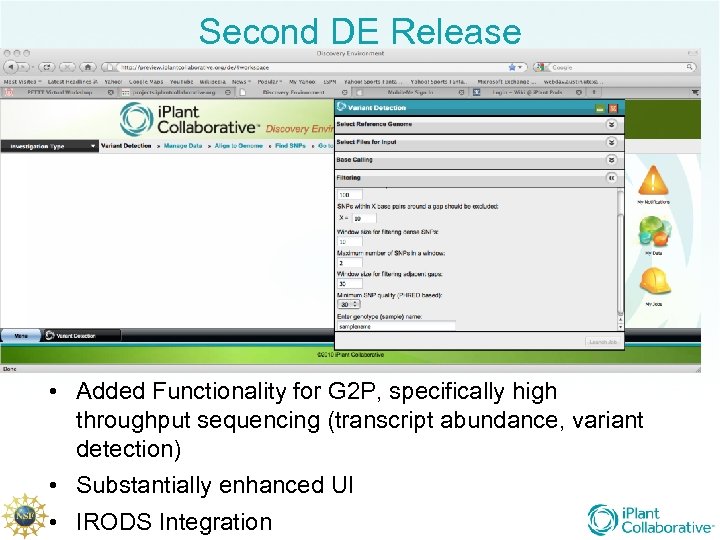
Second DE Release • Added Functionality for G 2 P, specifically high throughput sequencing (transcript abundance, variant detection) • Substantially enhanced UI • IRODS Integration

Portfolio of Activities • Maintaining a balance of “past, present, future” strategies – “Past”: make services, systems, and support available to existing bioinformatics projects, either to enhance them or simply make critical tools more widely available. – “Present” build the best bioinformatics software tools that today’s technologies can provide. – “Future” track emerging technologies, and where appropriate stimulate research into the creation and use of those technologies.

Portfolio of Activities • In a nutshell: – 12 Working groups in the two grand challenges, each of which is defining requirements for DE development. Each group not only has discussions that leads to final projects, but they also spawn prototyping efforts, tech eval projects, tool support projects, etc. – Services group: provide cycles, storage, hosting, etc. to users. – A comprehensive technology evaluation program to find, borrow, or build relevant technologies, headlined by the semantic web effort. – A number of ancillary projects related to grand challenges, i. e. APWEB, high throughput image analysis – The Core development/integration effort.
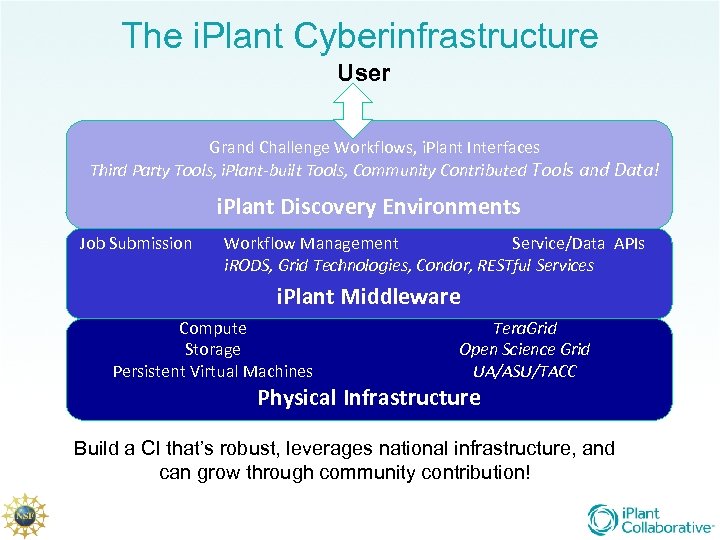
The i. Plant Cyberinfrastructure User Grand Challenge Workflows, i. Plant Interfaces Third Party Tools, i. Plant-built Tools, Community Contributed Tools and Data! i. Plant Discovery Environments Job Submission Workflow Management Service/Data APIs i. RODS, Grid Technologies, Condor, RESTful Services i. Plant Middleware Compute Storage Persistent Virtual Machines Tera. Grid Open Science Grid UA/ASU/TACC Physical Infrastructure Build a CI that’s robust, leverages national infrastructure, and can grow through community contribution!
Systems and Services • Provide access for problems like these on large scale systems • Provide the storage infrastructure for biological data (again, in support of existing projects) • Provide cloud style VM infrastructure for service hosting.

Existing Systems • We have made resources available to i. Plant users from a number of Tera. Grid and local systems – Ranger (TG/Large Scale Supercomputer) – Stampede (TACC/High Throughput) – Longhorn (TG/Remote Visualization and GPU) • The Contrast tool runs in production on Stampede; Tree. Viz on Longhorn • Several groups accessing these systems for real science now; command line only, but open for business!

Storage Services • We have also begun offering storage to a number of projects connected to the grand challenges in some way, as well as i. Plant internal. – IRODS interface – Corral at TACC, a local storage array at UA • Data arriving now for 1 KP project, Gates C 3/C 4 project, some labs starting to use… open for business.
Cloud Services • i. Plant is now offering “cloud” style hosting services. • Dynamically launch virtual servers hosted by i. Plant. • Still in prototype
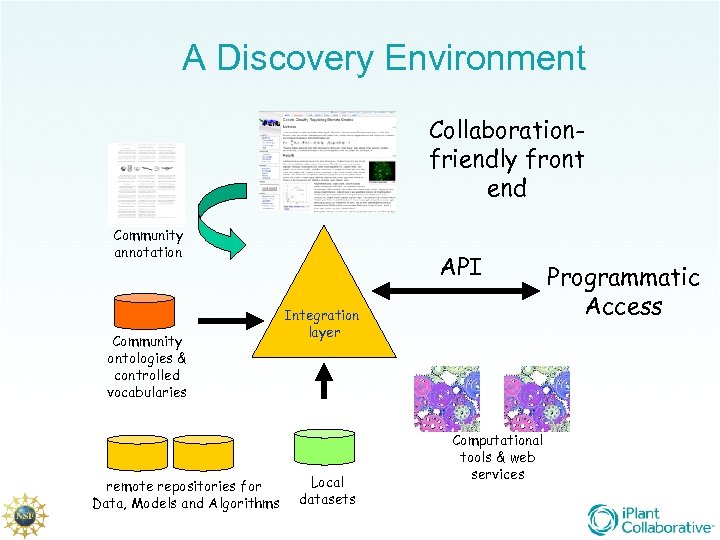
A Discovery Environment Collaborationfriendly front end Community annotation Community ontologies & controlled vocabularies remote repositories for Data, Models and Algorithms API Integration layer Local datasets Computational tools & web services Programmatic Access
Discovery Environment Releases • First release was March; Technology Preview – Tests of architecture, identity management, remote system integration, etc. – One supported workflow for IPTOL • Second release in June 2010 – Six workflows for IPG 2 P around high throughput sequencing – Integrated tree visualization tool – UI Refinements based on user feedback • Third release September 2010 – Basic Support for incorporation of 3 rd party tools – Enhanced collaboration features – Taxon name scrubber

The i. Plant Application Programmer Interface And What it Means to You
API – Why is it important? • The API is the Application Programmer Interface (it comes with an associated SDK, or software developer kit). • This is the way bioinformatics tools and data get integrated with i. Plant. • First pieces to be released late Sept/early October. The cardinal sin of API support: – Release lots of versions, each incompatible with the last. – Our approach: Incremental releases; each release will add new areas of functionality, not change old syntax – Initial support: Getting files in and out of the environment, running jobs.
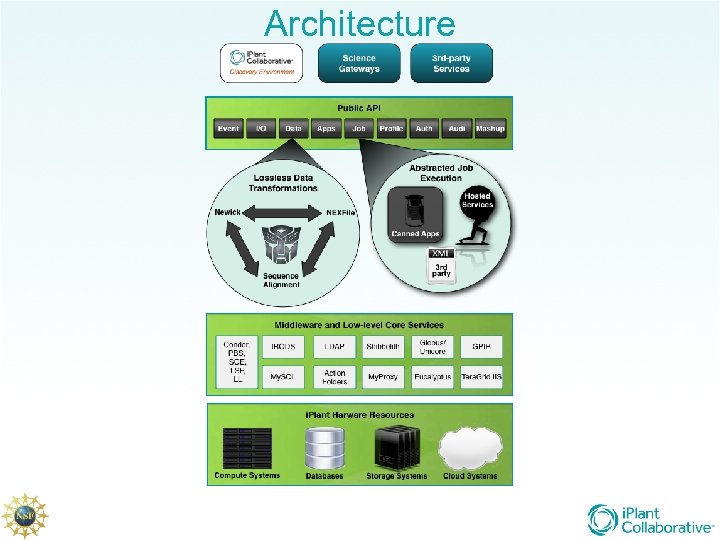
Architecture
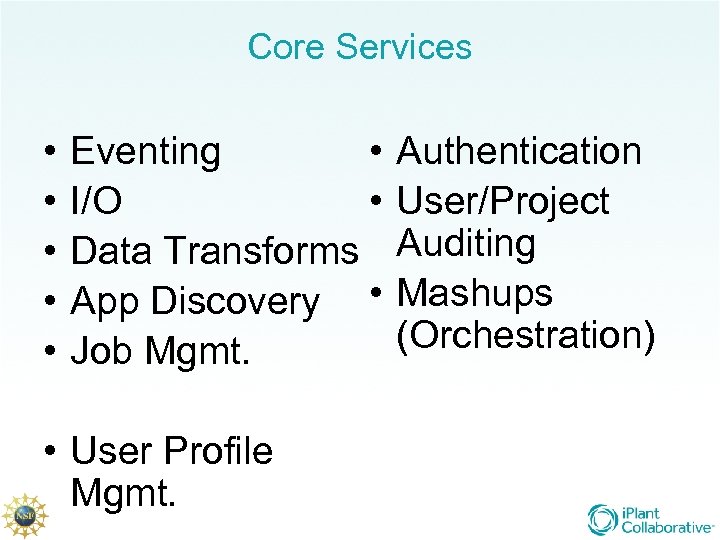
Core Services • • • Eventing • I/O Data Transforms App Discovery • Job Mgmt. • User Profile Mgmt. Authentication User/Project Auditing Mashups (Orchestration)
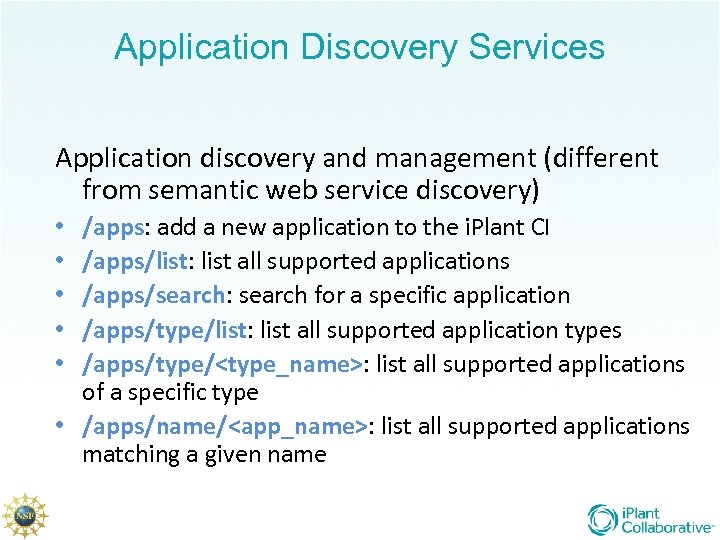
Application Discovery Services Application discovery and management (different from semantic web service discovery) /apps: add a new application to the i. Plant CI /apps/list: list all supported applications /apps/search: search for a specific application /apps/type/list: list all supported application types /apps/type/<type_name>: list all supported applications of a specific type • /apps/name/<app_name>: list all supported applications matching a given name • • •
API/DE Back End

Publishing Your Service Through i. Plant • Wrap your service in our API (or get us too). • Give us the package to deploy on our platforms (optional, but a good idea). • We register as a service, discoverable through app API. • Describe the user interface to our discovery environment (Graphical tool to build forms).

Using the API in your work *outside* the Discovery Environment • You need not come thru the DE to make use of a service. • Embed calls to the web service in your own code, or even from the command line. • For example, to get an output file from your Phylip run: https: //services. iplantcollaborative. org/contrast/file/get/(<id>) • While it is nice to do this by hand, the key thing is it can be *automated*.
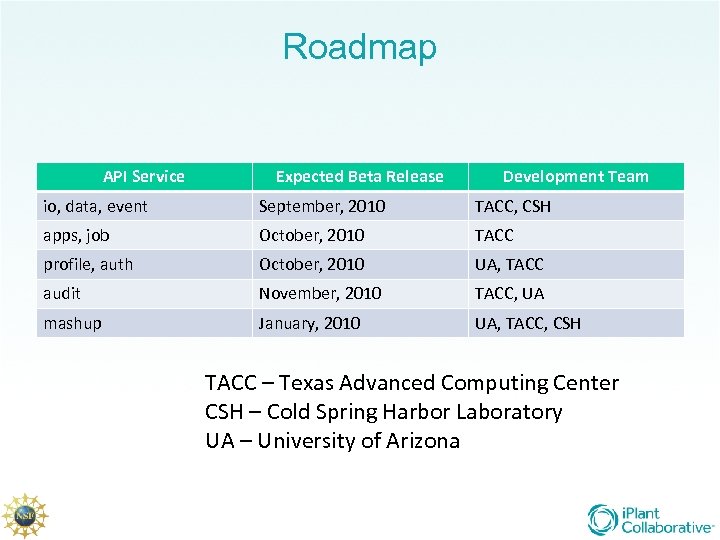
Roadmap API Service Expected Beta Release Development Team io, data, event September, 2010 TACC, CSH apps, job October, 2010 TACC profile, auth October, 2010 UA, TACC audit November, 2010 TACC, UA mashup January, 2010 UA, TACC, CSH TACC – Texas Advanced Computing Center CSH – Cold Spring Harbor Laboratory UA – University of Arizona

Technology Eval Activities • Largest investment in semantic web activities – Key for addressing the massive data integration challenges • Exploring alternate implementations of QTL mapping algorithms • Experimental Reproducability • Policy and Technology for Provenance Management • Evaluation of Hub. Zero, Workflow engines, numerous other tools

Deployment Strategy • Broadly, i. Plant CI deployment can be grouped into 3 categories: – Systems, services (middleware), and tools • In each category, there a couple of types of development/deployment activities. – Prototype, production • The transition from prototype to production can usually follow a relatively robust engineering schedule; prototyping less so.

So, what can I get from i. Plant Right NOW! • Tools: – Use the Discovery Environment to do transcript abundance, variant detection, trait evolution, or just store your stuff – Access to prototype tools for large scale tree visualization or very large tree building runs with neighbor joining or maximum likelihood. – Use My. Plant to find data and colleagues working on related species – Use DNASubway to do genome annotation and train your students.

So, what can I get from i. Plant Right NOW! • Systems/Services: – Request a repository to provide command line or Web. DAV access to large scale datasets on high integrity storage systems – Get command line access to the most powerful computing and visualization systems in the world. – Use the i. Plant Cloud to host your web application in a virtual machine.

So, what can I get from i. Plant SOON • Services: – Use the API to embed access to i. Plant tools, systems, and data repositories in your own scripts and workflows. – Submit your bioinformatics tool to be registered as an i. Plant service (run on large platforms, available to others thru API), or make your web service discoverable thru i. Plant. A little later: Have your tool incorporated in the i. Plant DE with it’s own graphical interface. • Tools: more coming on line steadily

Collaborations • More than 80 faculty at 45 institutions involved in working groups. • Gates Integrated Breeding Platform • Gates C 3/C 4 photosynthesis project • 1 KP thousand plant transcriptome project • Nascent “National Virtual Herbaria”, many existing herbaria.

Discussion (See demo clips at http: //iplantcollaborative. org/videos

CI Master Project List • DE • Ingest pipeline (Phlawd) • API • DNA Subway • Semantic Web • Brachy. Bio • GLM • Drop. Box • GLM – GPU • Cloud service • My. Plant • Storage repositories • Ra. XML • Analytics pipeline • NINJA • Visualization explorations • Experimental Reproducability • Workflow tools analysis • Image management pipeline • APWEB refit • Large Scale Tree Visualization • Name resolution service
DNA Subway

My Plant • Social networking for plant biologists • Organized by clade • Used to organize the data collection for the “big tree”

Scope: What i. Plant won’t do • i. Plant is not a funding agency – A large grant shouldn’t become a bunch of small grants • i. Plant does not fund data collection • i. Plant will (probably) not continue funding for <favorite tool x> whose funding is ending. • i. Plant will not seek to replace all online data repositories • i. Plant will not *impose* standards on the community.

Scope: What i. Plant *will* do • Provide storage, computation, hosting, and lots of programmer effort to support grand challenge efforts. • Work with the community to support and develop standards • Provide forums to discuss the role and design of CI in plant science • Help organize the community to collect data • Provide appropriate funding for time spent helping us design and test the CI
Experimental Systems • We are experimenting with some newer technologies to plug gaps in the existing lineup for demonstrated needs (also leveraging some other funding) – New model for shared memory (Scale. MP cluster to be deployed soon) Will support whole-genome assembly – “Cloud Storage” models to reduce archive cost, increase capacity (HDFS system on commodity cluster to be deployed this quarter) Will also support Hadoop data processing
Deployment Timelines Summary • Systems: – Production systems (HPC, storage, throughput, visualization) available *now* and in use. – Experimental systems (simulated shared memory, cloud storage) coming up in prototype stage. • Services: – Web service API to incorporate 3 rd party tools prototyping now, public releases in Q 3. • Tools – A number of prototypes available now, many underway – Contrast workflow, Variant detection, Transcript quantification all released now.

The i. Plant CI • Engagement with the CI Community to leverage best practice and new research • Unprecedented engagement with the user community to drive requirements • A single CI for all plant scientists, with customized discovery environments to meet grand challenges • An exemplar virtual organization for modern computational science • A Foundation of Computational and Storage Capability • Open source principles, commercial quality development process
9b8aebba0916402af1e7f79f6be3f710.ppt