a2d0d82654146a7f97539e8f46f733a1.ppt
- Количество слайдов: 36
The first Korean Genome Sequence analysis using Bioinformatics Jong Bhak 20091120 jongbhak@yahoo. com Theragen Inc. 테라젠(주)

Acknowledgement • Gacheon Med. School. LCDi( Lee Gilya Cancer Diabetes Inst. ) • 김성진박사님, 안성민박사님 • 키스티 • 정민중박사님 • Theragen Inc. (테라젠 (주))
Human Genome 3 GB • 6, 000 km (Seoul Moscow: 6, 600 km) • SF NY (4, 100 km) • London Boson (5, 300 km)

Current Status & Prediction • Genome era has arrived in 2007 ~ 2008 • Bioinformatics is becoming “industrial” in 2008 • The Bio. Revolution started and revolutionizing – – the bioresearch, medical, healthcare, industrial, and information tech. by 2016
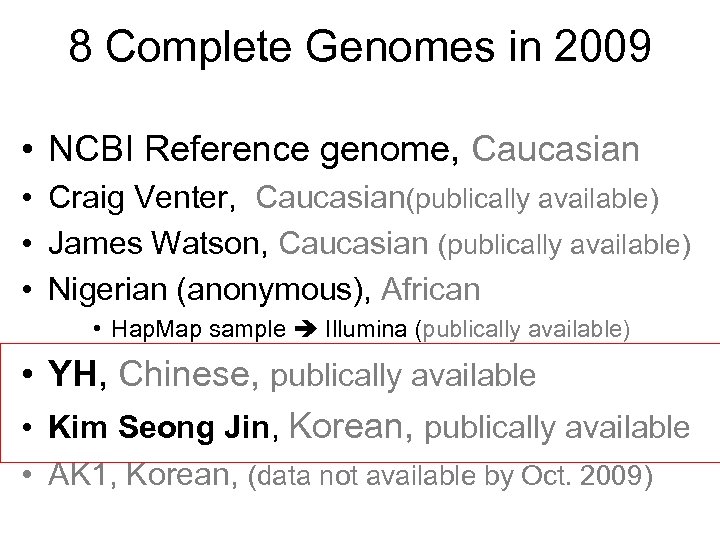
8 Complete Genomes in 2009 • NCBI Reference genome, Caucasian • Craig Venter, Caucasian(publically available) • James Watson, Caucasian (publically available) • Nigerian (anonymous), African • Hap. Map sample Illumina (publically available) • YH, Chinese, publically available • Kim Seong Jin, Korean, publically available • AK 1, Korean, (data not available by Oct. 2009)
DNA sequencing • First genome sequencing: 1977 Sanger method – Phi X 174 – Mitochondrial genome (1981) • 1998: Theoretically it takes one day to sequence a human genome (Church Lab, Harvard) • Polony based (Church, Knome. Inc) • 454 • Sollexa • Now: Over 2 GB per experiment Jong Bhak, under openfree Bio. License

Cost • • NCBI reference genome: 3, 000 million USD Craig Venter: 100 million USD James Watson: 1 million USD YH Chinese: 0. 5 million USD Nigerian African: 0. 25 million USD (Illumina) Kim Sung Jin: 0. 25 million USD Complete Genomics: 0. 005 million USD • 2010: 0. 001 million USD • 2012: $100 USD?
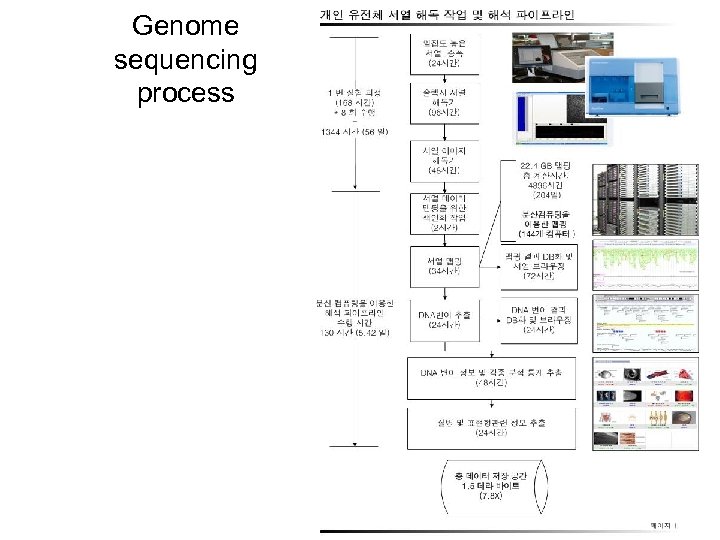
Genome sequencing process

Genomics era? Full genome sequencing Full genomics Individual sequencing cost can be $1000 or $100 by the year 2013 However,

Genomics era? However, - Useful Genomics person can still cost $10, 000 or more $1000 genomics Personal Genome $0 Genomics an openfree genomics project
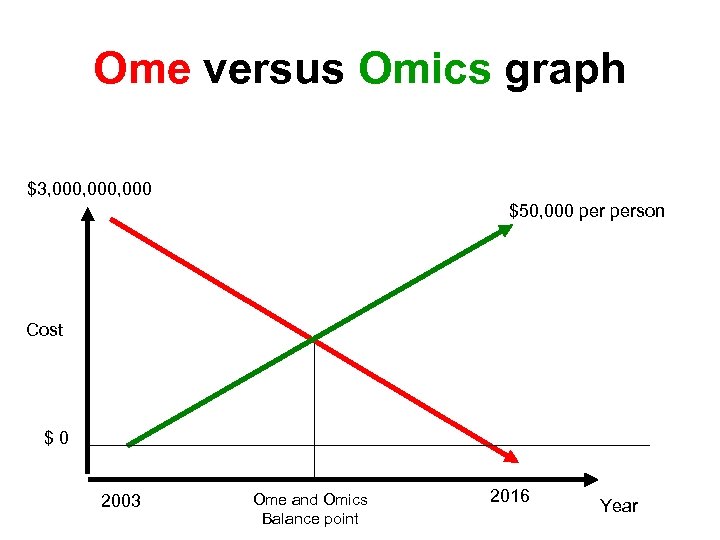
Ome versus Omics graph $3, 000, 000 $50, 000 person Cost $0 2003 Ome and Omics Balance point 2016 Year
The most important aspect of Genomics: Variomics • Personal genome comparison is now possible – Personal comparative genomics Variomics Genomics Jong Bhak, under openfree Bio. License

A large pool of variation information – Provide the map for global human genome(s) project saves money. – Provide association studies on all ethnic groups and individuals for phenotyping. – Extract disease association information. – Mapping everyone’s distance to each other. Human diversity. – Provide the public with an openfree personal variome analysis package.

The Korean Genome
The first Korean Genome (SJK) • First analyzed by Gacheon medical school LCDI and KOBIC, KRIBB in 2008 (Joint effort among LCDI, KOBIC, and 국가참조 표준센터) • First annotated and made public on 4 th Dec. 2008 (through web and ftp) • To be used as the first National Reference Genome • SNP, CNV, indels were analysed • Automated phenotypic association study was done • Non-syn. Analysis • Phylogenetic study of mt. DNA, Y Chr And autosomes showed Korean relationship to Chinese and Japanese. • First intra-Asian genome comparison (Chinese and Korean) • Analyzed at: 7. 8, 17. 3, 23. 5 and 28 x folds • By Jan. 23. 5 fold sequenced analyzed • Openfreely Available from: http: //koreagenome. org
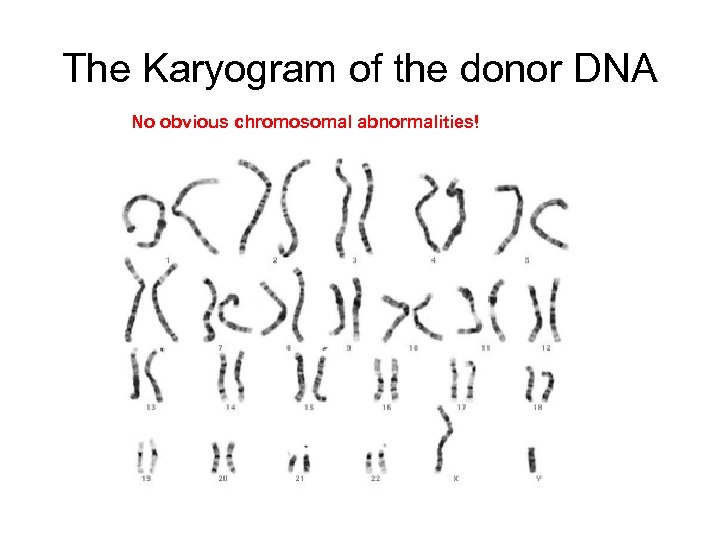
The Karyogram of the donor DNA No obvious chromosomal abnormalities!
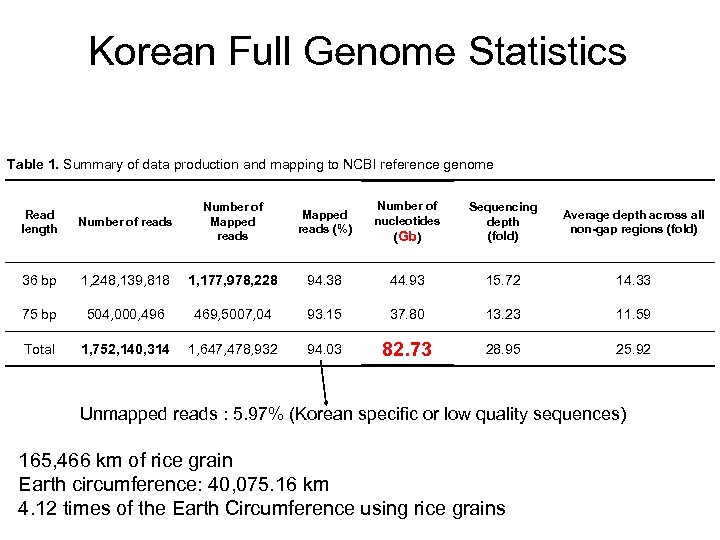
Korean Full Genome Statistics Table 1. Summary of data production and mapping to NCBI reference genome Number of reads Number of Mapped reads (%) Number of nucleotides (Gb) Sequencing depth (fold) Average depth across all non-gap regions (fold) 36 bp 1, 248, 139, 818 1, 177, 978, 228 94. 38 44. 93 15. 72 14. 33 75 bp 504, 000, 496 469, 5007, 04 93. 15 37. 80 13. 23 11. 59 Total 1, 752, 140, 314 1, 647, 478, 932 94. 03 82. 73 28. 95 25. 92 Read length Unmapped reads : 5. 97% (Korean specific or low quality sequences) 165, 466 km of rice grain Earth circumference: 40, 075. 16 km 4. 12 times of the Earth Circumference using rice grains
Variation and Variomics
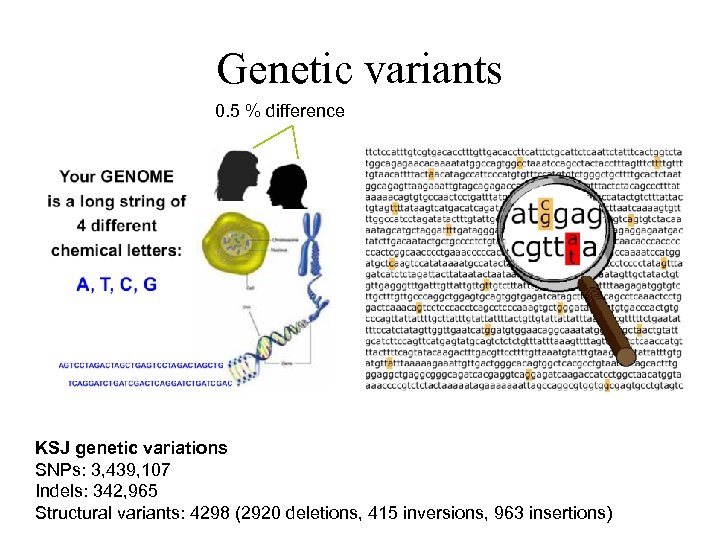
Genetic variants 0. 5 % difference KSJ genetic variations SNPs: 3, 439, 107 Indels: 342, 965 Structural variants: 4298 (2920 deletions, 415 inversions, 963 insertions)
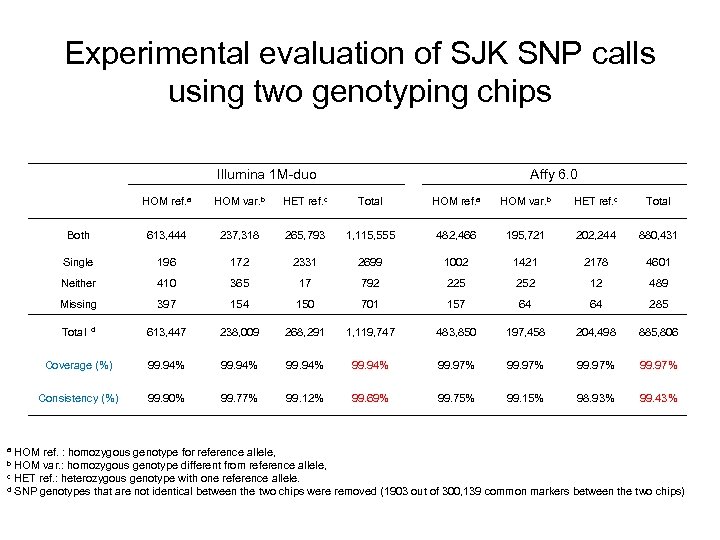
Experimental evaluation of SJK SNP calls using two genotyping chips Illumina 1 M-duo Affy 6. 0 HOM ref. a HOM var. b HET ref. c Total Both 613, 444 237, 318 265, 793 1, 115, 555 482, 466 195, 721 202, 244 880, 431 Single 196 172 2331 2699 1002 1421 2178 4601 Neither 410 365 17 792 225 252 12 489 Missing 397 154 150 701 157 64 64 285 613, 447 238, 009 268, 291 1, 119, 747 483, 850 197, 458 204, 498 885, 806 Coverage (%) 99. 94% 99. 97% Consistency (%) 99. 90% 99. 77% 99. 12% 99. 69% 99. 75% 99. 15% 98. 93% 99. 43% Total d HOM ref. : homozygous genotype for reference allele, HOM var. : homozygous genotype different from reference allele, c HET ref. : heterozygous genotype with one reference allele. d SNP genotypes that are not identical between the two chips were removed (1903 out of 300, 139 common markers between the two chips) a b
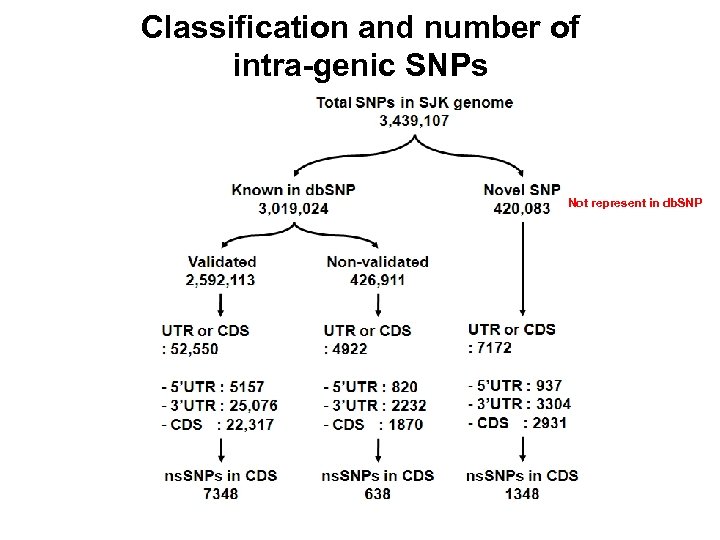
Classification and number of intra-genic SNPs Not represent in db. SNP
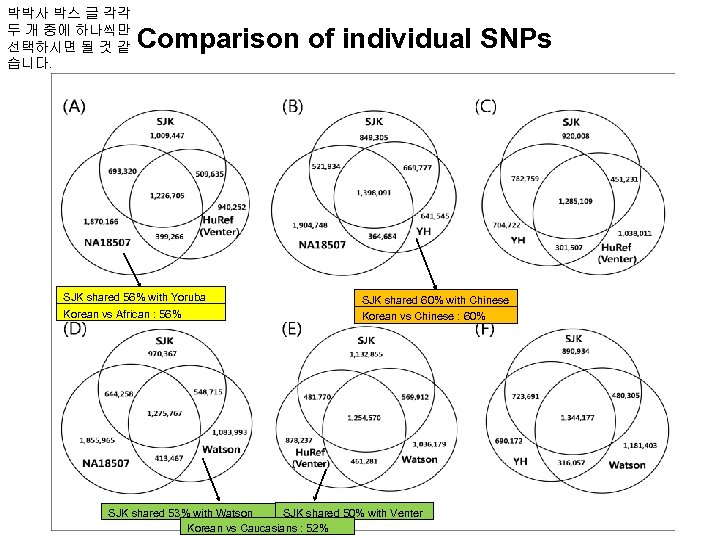
박박사 박스 글 각각 두 개 중에 하나씩만 선택하시면 될 것 같 습니다. Comparison of individual SNPs SJK shared 56% with Yoruba SJK shared 60% with Chinese Korean vs African : 56% Korean vs Chinese : 60% SJK shared 50% with Venter SJK shared 53% with Watson Korean vs Caucasians : 52%
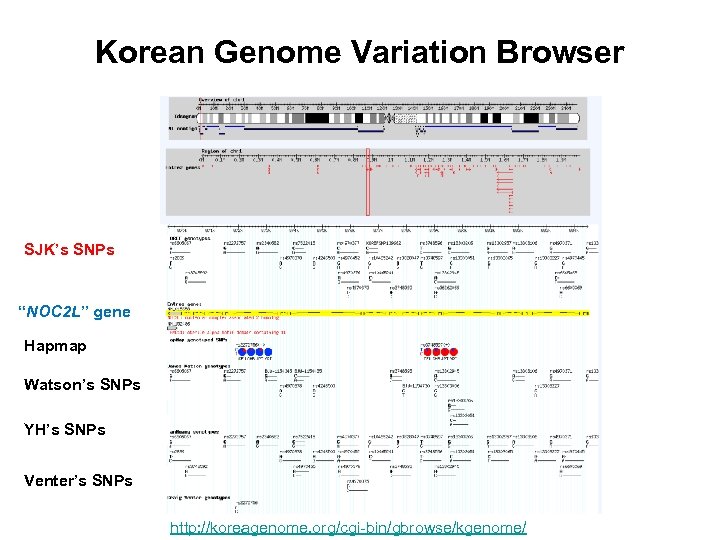
Korean Genome Variation Browser SJK’s SNPs “NOC 2 L” gene Hapmap Watson’s SNPs YH’s SNPs Venter’s SNPs http: //koreagenome. org/cgi-bin/gbrowse/kgenome/
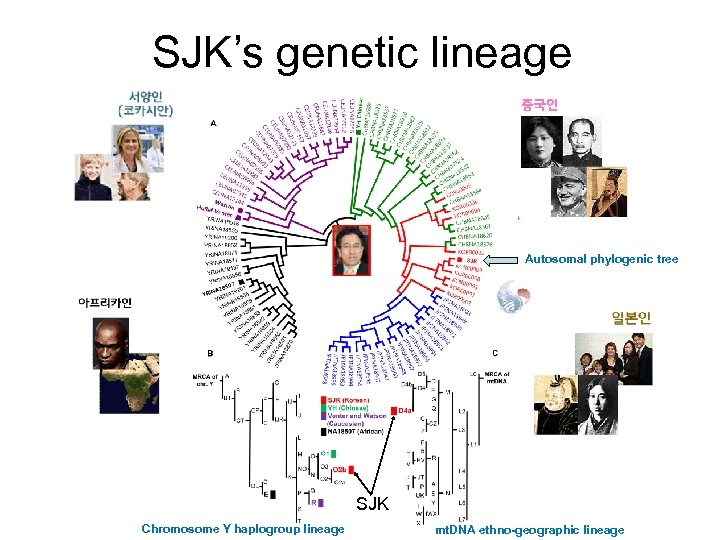
SJK’s genetic lineage Autosomal phylogenic tree SJK Chromosome Y haplogroup lineage mt. DNA ethno-geographic lineage
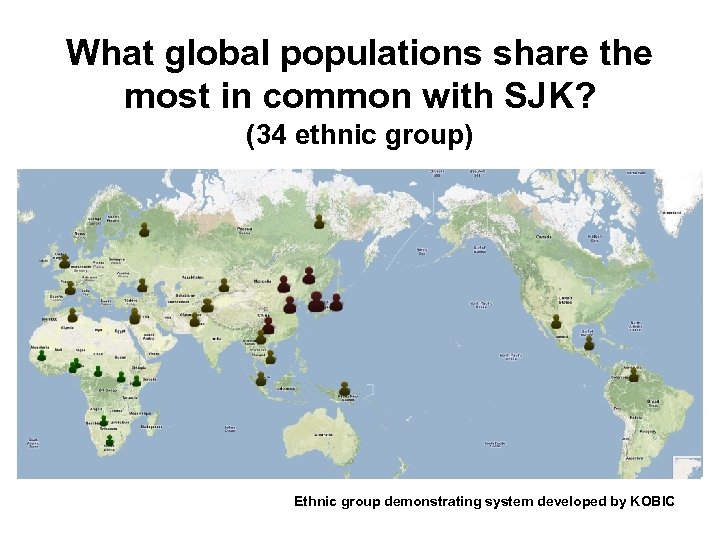
What global populations share the most in common with SJK? (34 ethnic group) Ethnic group demonstrating system developed by KOBIC
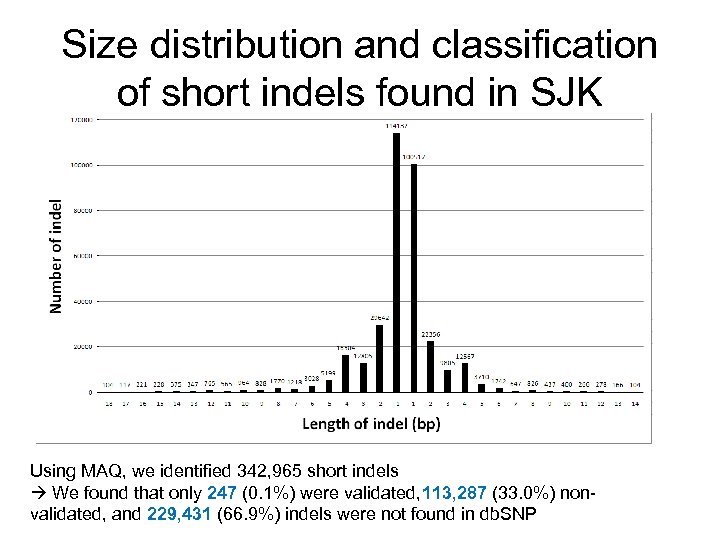
Size distribution and classification of short indels found in SJK Using MAQ, we identified 342, 965 short indels We found that only 247 (0. 1%) were validated, 113, 287 (33. 0%) nonvalidated, and 229, 431 (66. 9%) indels were not found in db. SNP
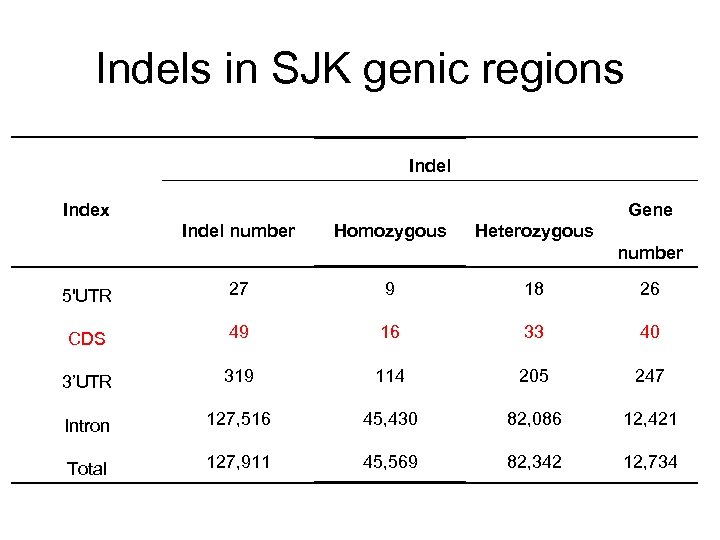
Indels in SJK genic regions Indel Gene Index Indel number Homozygous Heterozygous number 5'UTR 27 9 18 26 CDS 49 16 33 40 3’UTR 319 114 205 247 Intron 127, 516 45, 430 82, 086 12, 421 Total 127, 911 45, 569 82, 342 12, 734
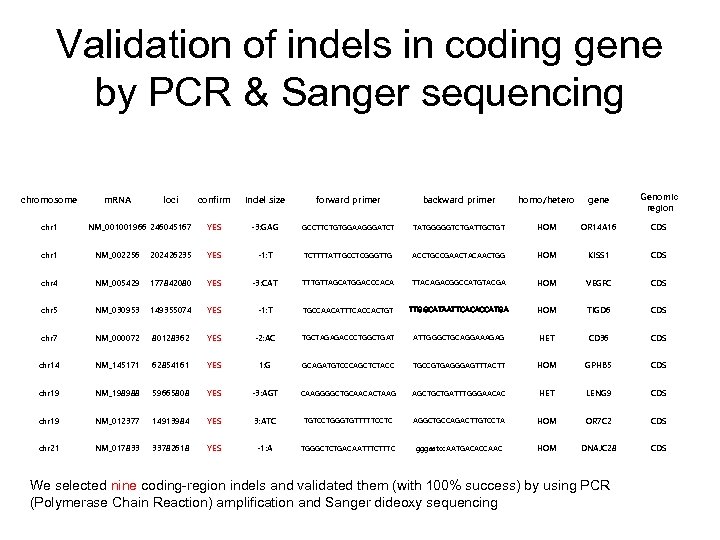
Validation of indels in coding gene by PCR & Sanger sequencing chromosome chr 1 m. RNA loci NM_001001966 246045167 confirm Indel size forward primer backward primer homo/hetero gene Genomic region YES -3: GAG GCCTTCTGTGGAAGGGATCT TATGGGGGTCTGATTGCTGT HOM OR 14 A 16 CDS chr 1 NM_002256 202426235 YES -1: T TCTTTTATTGCCTCGGGTTG ACCTGCCGAACTACAACTGG HOM KISS 1 CDS chr 4 NM_005429 177842080 YES -3: CAT TTTGTTAGCATGGACCCACA TTACAGACGGCCATGTACGA HOM VEGFC CDS chr 5 NM_030953 149355074 YES -1: T TGCCAACATTTCACCACTGT TTGGCATAATTCACACCATGA HOM TIGD 6 CDS chr 7 NM_000072 80128362 YES -2: AC TGCTAGAGACCCTGGCTGAT ATTGGGCTGCAGGAAAGAG HET CD 36 CDS chr 14 NM_145171 62854161 YES 1: G GCAGATGTCCCAGCTCTACC TGCCGTGAGGGAGTTTACTT HOM GPHB 5 CDS chr 19 NM_198988 59665808 YES -3: AGT CAAGGGGCTGCAACACTAAG AGCTGCTGATTTGGGAACAC HET LENG 9 CDS chr 19 NM_012377 14913984 YES 3: ATC TGTCCTGGGTGTTTTTCCTC AGGCTGCCAGACTTGTCCTA HOM OR 7 C 2 CDS chr 21 NM_017833 33782618 YES -1: A TGGGCTCTGACAATTTC gggaatcc. AATGACACCAAC HOM DNAJC 28 CDS We selected nine coding-region indels and validated them (with 100% success) by using PCR (Polymerase Chain Reaction) amplification and Sanger dideoxy sequencing
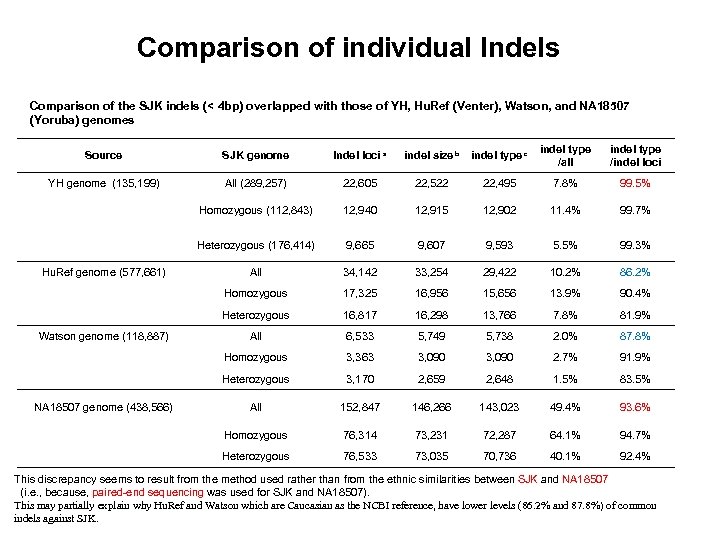
Comparison of individual Indels Comparison of the SJK indels (< 4 bp) overlapped with those of YH, Hu. Ref (Venter), Watson, and NA 18507 (Yoruba) genomes Source SJK genome Indel loci a indel size b indel type c indel type /all indel type /indel loci YH genome (135, 199) All (289, 257) 22, 605 22, 522 22, 495 7. 8% 99. 5% Homozygous (112, 843) 12, 940 12, 915 12, 902 11. 4% 99. 7% Heterozygous (176, 414) 9, 665 9, 607 9, 593 5. 5% 99. 3% Hu. Ref genome (577, 661) All 34, 142 33, 254 29, 422 10. 2% 86. 2% Homozygous 17, 325 16, 956 15, 656 13. 9% 90. 4% Heterozygous 16, 817 16, 298 13, 766 7. 8% 81. 9% Watson genome (118, 887) All 6, 533 5, 749 5, 738 2. 0% 87. 8% Homozygous 3, 363 3, 090 2. 7% 91. 9% Heterozygous 3, 170 2, 659 2, 648 1. 5% 83. 5% NA 18507 genome (438, 566) All 152, 847 146, 266 143, 023 49. 4% 93. 6% Homozygous 76, 314 73, 231 72, 287 64. 1% 94. 7% Heterozygous 76, 533 73, 035 70, 736 40. 1% 92. 4% This discrepancy seems to result from the method used rather than from the ethnic similarities between SJK and NA 18507 (i. e. , because, paired-end sequencing was used for SJK and NA 18507). This may partially explain why Hu. Ref and Watson which are Caucasian as the NCBI reference, have lower levels (86. 2% and 87. 8%) of common indels against SJK.
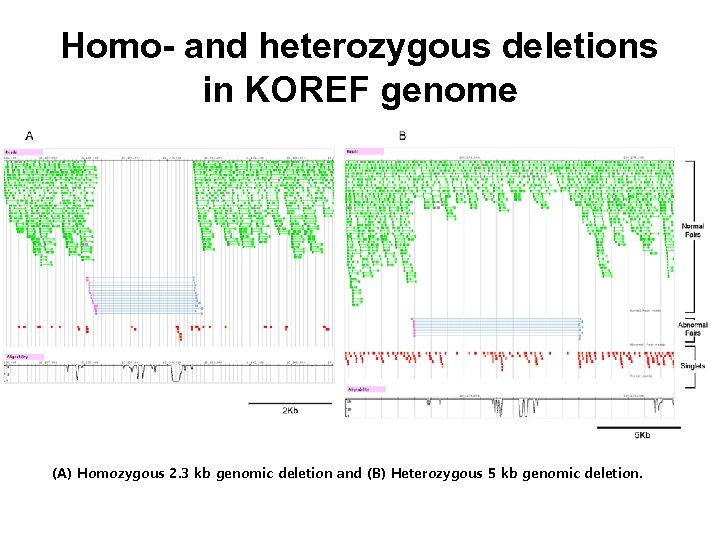
Homo- and heterozygous deletions in KOREF genome (A) Homozygous 2. 3 kb genomic deletion and (B) Heterozygous 5 kb genomic deletion.
Detection and identification of structural variants • We found structural variants by using paired-end reads. 1. 2920 deletions (100 bp ~ 100 kb) 2. 415 inversions (100 bp ~ 100 kb) 3. 963 insertions (175 bp ~ 250 bp) • We found deletion SVs in 21 coding genes. All heterozygous deletions
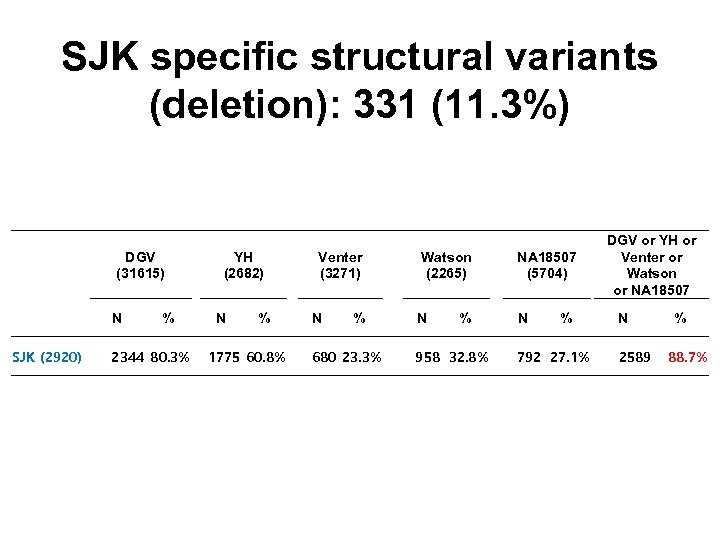
SJK specific structural variants (deletion): 331 (11. 3%) DGV (31615) YH (2682) Venter (3271) N SJK (2920) 2344 80. 3% 1775 60. 8% 680 23. 3% % N % Watson (2265) N % NA 18507 (5704) DGV or YH or Venter or Watson or NA 18507 N % 958 32. 8% 792 27. 1% 2589 % 88. 7%
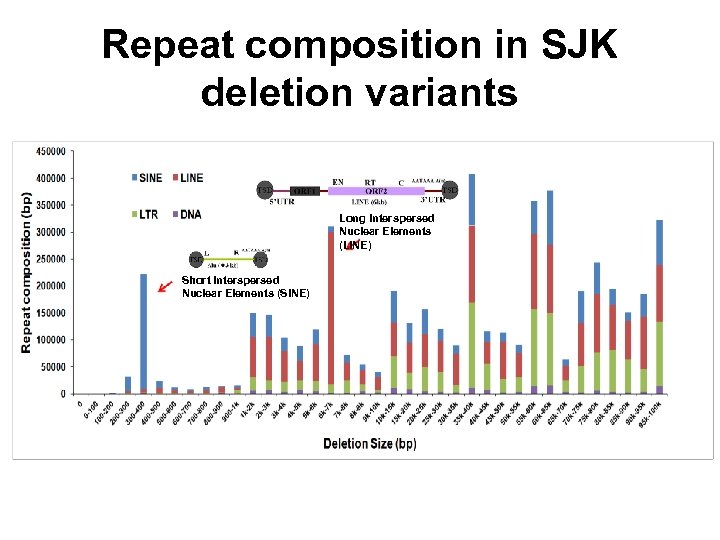
Repeat composition in SJK deletion variants Long Interspersed Nuclear Elements (LINE) Short Interspersed Nuclear Elements (SINE)
Genomics & Bioinformatics in Theragen • • Genomics and Bioinformatics company Marker discovery Drug Target & Drug screening Personalized, Preventive, Predictive medicine • Genomics experiment team + Bioinformatics team • 연구소: 광교 테크노 밸리, 차세대 융합기술원 2층, 동수원 IC
Genome Information Data Center • • • Genome information data center Much experience in handling biodata Top level of bioinformatics/DB handling in the world International network Experience in maintaining large clusters of CPUs and storage
a2d0d82654146a7f97539e8f46f733a1.ppt