f531b92f1354c0836868dffdefc5cfd3.ppt
- Количество слайдов: 35
The Eco. Cyc and Meta. Cyc Pathway/Genome Databases Peter D. Karp, Ph. D. Bioinformatics Research Group SRI International pkarp@ai. sri. com http: //www. ai. sri. com/pkarp/ http: //Eco. Cyc. org/

SRI International Bioinformatics Overview l Motivations and terminology l Pathway/genome databases l Bio. Cyc collection l Eco. Cyc, Meta. Cyc l Pathway Tools software l Bioinformatics Database Warehouse project
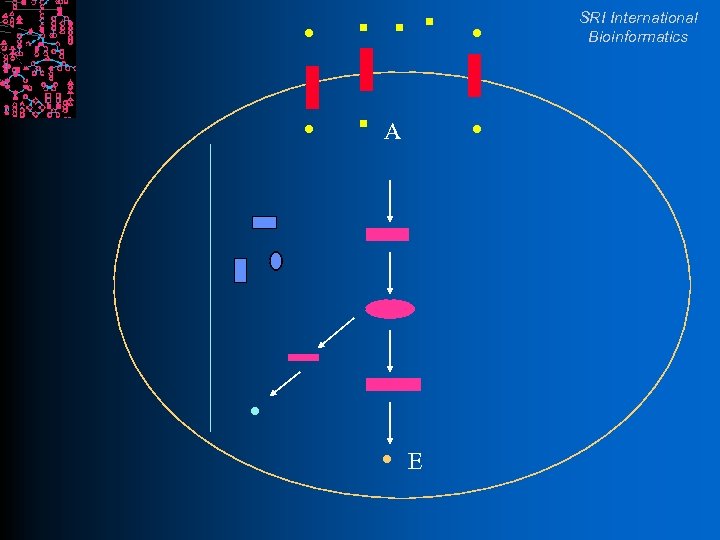
SRI International Bioinformatics A E

What to do When Theories Become Larger than Minds can Grasp? l l SRI International Bioinformatics Example: E. coli metabolic network l 160 pathways involving 744 reactions and 791 substrates Example: E. coli genetic network l Control by 97 transcription factors of 1174 genes in 630 transcription units Past solutions: l Partition theories across multiple minds l Encode theories in natural-language text We cannot compute with theories in those forms l Evaluate theories for consistency with new data: microarrays l Refine theories with respect to new data l Compare theories describing different organisms

Solution: Biological Knowledge Bases SRI International Bioinformatics l Store biological knowledge and theories in computers in a declarative form l Amenable to computational analysis and generative user interfaces l Establish ongoing efforts to curate (maintain, refine, embellish) these knowledge bases l Accepted to store data in computers, but not knowledge l Such knowledge bases are an integral part of the scientific enterprise
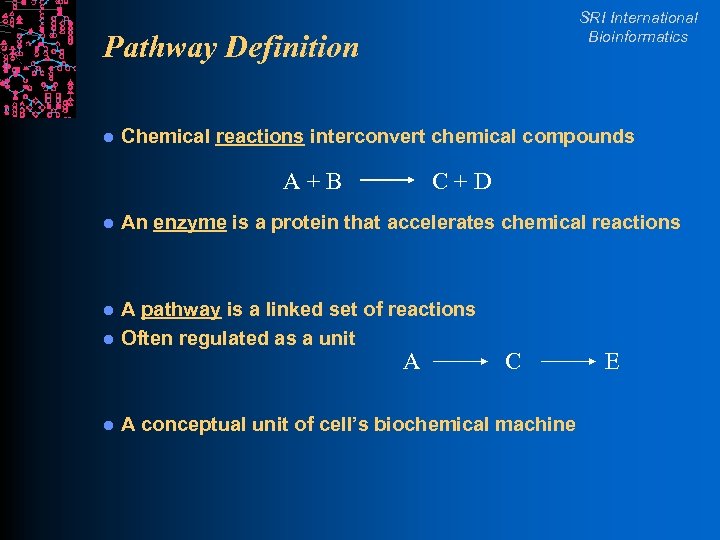
SRI International Bioinformatics Pathway Definition l Chemical reactions interconvert chemical compounds A+B C+D l An enzyme is a protein that accelerates chemical reactions l l A pathway is a linked set of reactions Often regulated as a unit l A conceptual unit of cell’s biochemical machine A C E
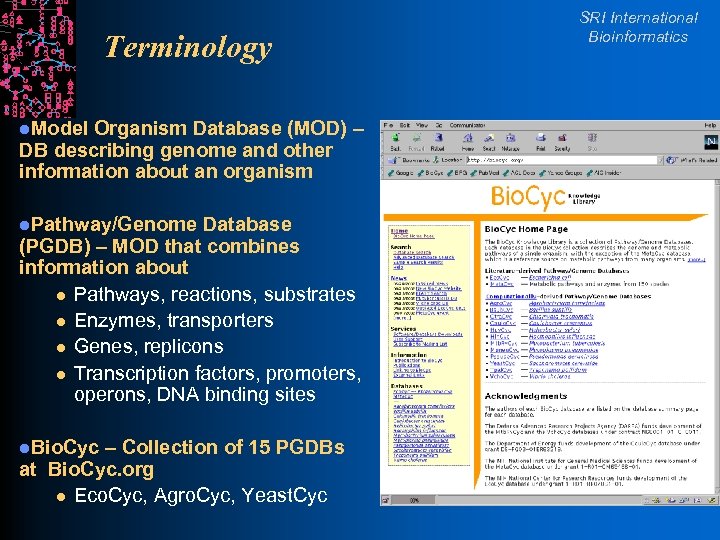
Terminology l. Model Organism Database (MOD) – DB describing genome and other information about an organism l. Pathway/Genome Database (PGDB) – MOD that combines information about l Pathways, reactions, substrates l Enzymes, transporters l Genes, replicons l Transcription factors, promoters, operons, DNA binding sites l. Bio. Cyc – Collection of 15 PGDBs at Bio. Cyc. org l Eco. Cyc, Agro. Cyc, Yeast. Cyc SRI International Bioinformatics
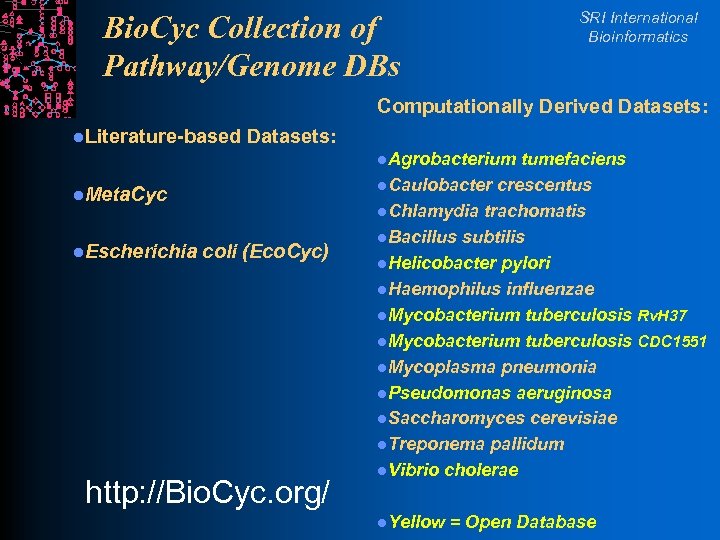
SRI International Bioinformatics Bio. Cyc Collection of Pathway/Genome DBs Computationally Derived Datasets: l. Literature-based Datasets: l. Agrobacterium l. Meta. Cyc l. Escherichia coli (Eco. Cyc) http: //Bio. Cyc. org/ tumefaciens l. Caulobacter crescentus l. Chlamydia trachomatis l. Bacillus subtilis l. Helicobacter pylori l. Haemophilus influenzae l. Mycobacterium tuberculosis Rv. H 37 l. Mycobacterium tuberculosis CDC 1551 l. Mycoplasma pneumonia l. Pseudomonas aeruginosa l. Saccharomyces cerevisiae l. Treponema pallidum l. Vibrio cholerae l. Yellow = Open Database
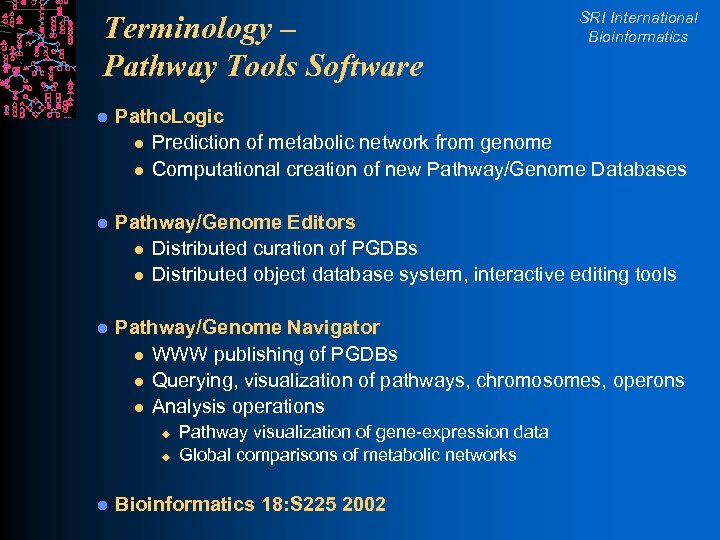
Terminology – Pathway Tools Software SRI International Bioinformatics l Patho. Logic l Prediction of metabolic network from genome l Computational creation of new Pathway/Genome Databases l Pathway/Genome Editors l Distributed curation of PGDBs l Distributed object database system, interactive editing tools l Pathway/Genome Navigator l WWW publishing of PGDBs l Querying, visualization of pathways, chromosomes, operons l Analysis operations u u l Pathway visualization of gene-expression data Global comparisons of metabolic networks Bioinformatics 18: S 225 2002

Pathway Tools Algorithms l. Query, visualization and editing tools for these datatypes: l. Full Metabolic Map l Paint gene expression data on metabolic network; compare metabolic networks l. Pathways l Pathway prediction l. Reactions l Balance checker l. Compounds l Chemical substructure comparison l. Enzymes, Transporters, Transcription Factors l. Genes: Blast search l. Chromosomes l. Operons SRI International Bioinformatics
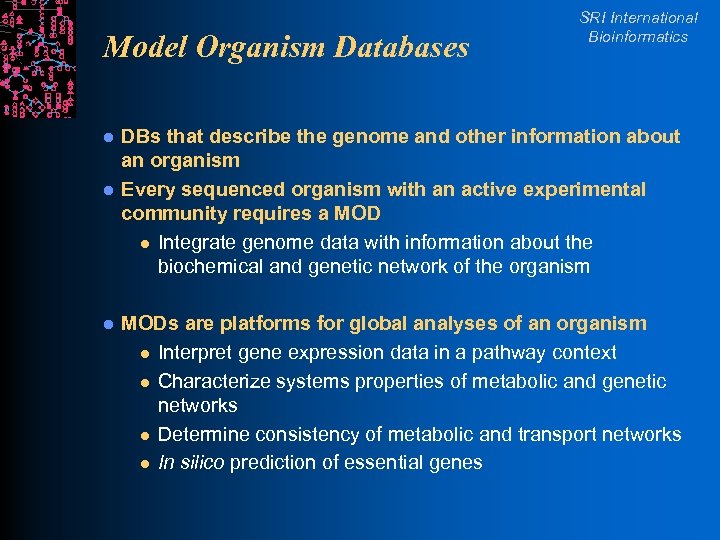
Model Organism Databases l l l SRI International Bioinformatics DBs that describe the genome and other information about an organism Every sequenced organism with an active experimental community requires a MOD l Integrate genome data with information about the biochemical and genetic network of the organism MODs are platforms for global analyses of an organism l Interpret gene expression data in a pathway context l Characterize systems properties of metabolic and genetic networks l Determine consistency of metabolic and transport networks l In silico prediction of essential genes

Eco. Cyc Project – Eco. Cyc. org SRI International Bioinformatics l E. coli Encyclopedia l Model-Organism Database for E. coli l Computational symbolic theory of E. coli l Electronic review article for E. coli – over 3500 literature citations l Tracks the evolving annotation of the E. coli genome l Collaborative development via Internet l Karp (SRI) -- Bioinformatics architect l John Ingraham -- Advisor l (SRI) Metabolic pathways l Saier (UCSD) and Paulsen (TIGR)-- Transport l Collado (UNAM)-- Regulation of gene expression l Database content: 18, 000 objects
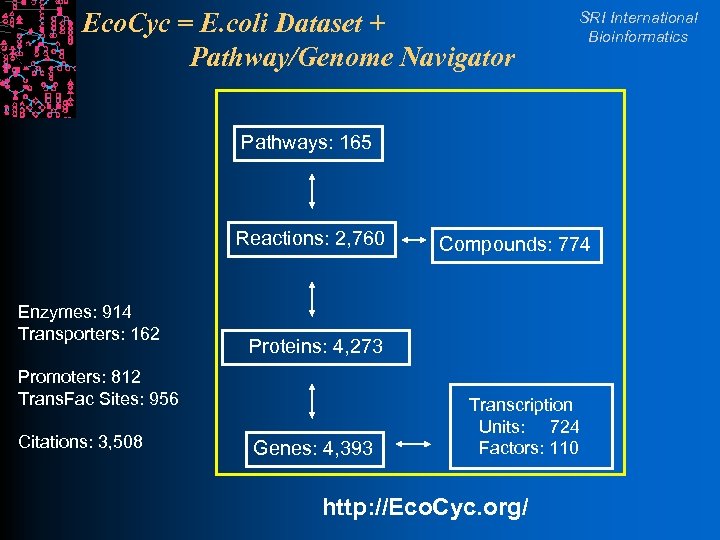
Eco. Cyc = E. coli Dataset + Pathway/Genome Navigator SRI International Bioinformatics Pathways: 165 Reactions: 2, 760 Enzymes: 914 Transporters: 162 Proteins: 4, 273 Promoters: 812 Trans. Fac Sites: 956 Citations: 3, 508 Compounds: 774 Genes: 4, 393 Transcription Units: 724 Factors: 110 http: //Eco. Cyc. org/

Eco. Cyc Procedures l All SRI International Bioinformatics DB updates by 5 staff curators l Information gathered from biomedical literature l Corrections solicited from E. coli researchers l Review-level database l Four releases per year l Available through WWW site, as data files, as downloadable application l Quality assurance of data and software: l Evaluate database consistency constraints l Perform element balancing of reactions l Run other checking programs l Display every DB object
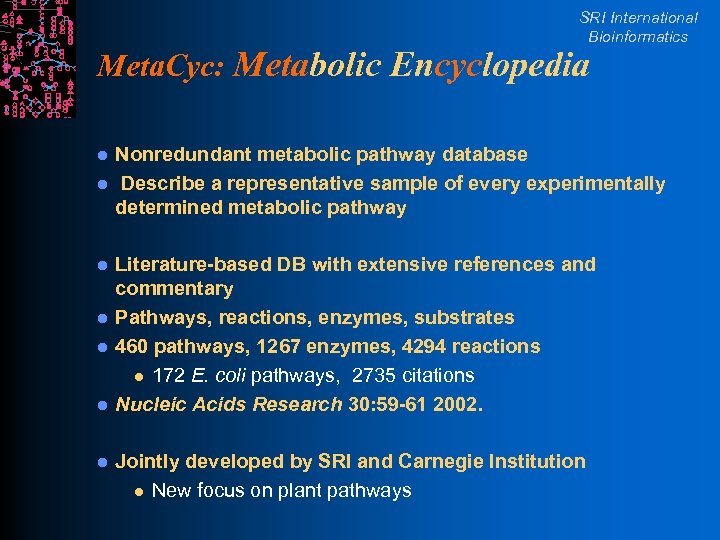
SRI International Bioinformatics Meta. Cyc: Metabolic Encyclopedia l l l l Nonredundant metabolic pathway database Describe a representative sample of every experimentally determined metabolic pathway Literature-based DB with extensive references and commentary Pathways, reactions, enzymes, substrates 460 pathways, 1267 enzymes, 4294 reactions l 172 E. coli pathways, 2735 citations Nucleic Acids Research 30: 59 -61 2002. Jointly developed by SRI and Carnegie Institution l New focus on plant pathways
Family of Pathway/Genome Databases Meta. Cyc SRI International Bioinformatics

SRI International Bioinformatics Pathway Tools Implementation Details l Allegro Common Lisp l Sun and PC platforms l Ocelot object database l 250, 000 lines of code l Lisp-based WWW server at Bio. Cyc. org l Manages 15 PGDBs
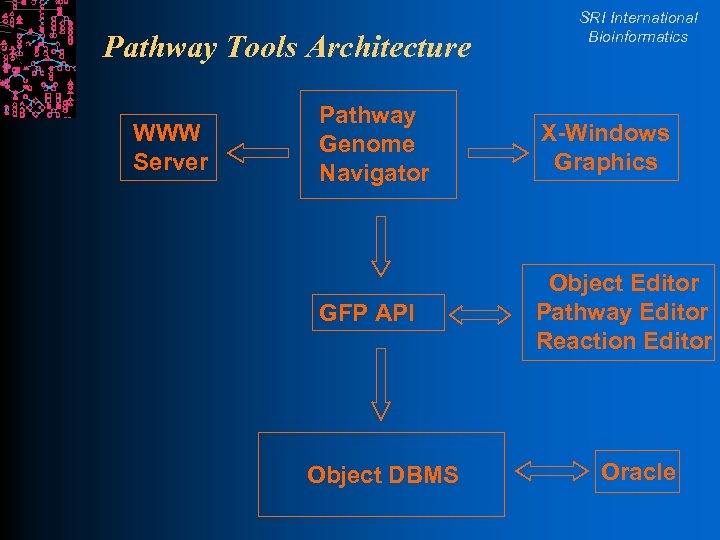
Pathway Tools Architecture WWW Server SRI International Bioinformatics Pathway Genome Navigator X-Windows Graphics GFP API Object Editor Pathway Editor Reaction Editor Object DBMS Oracle
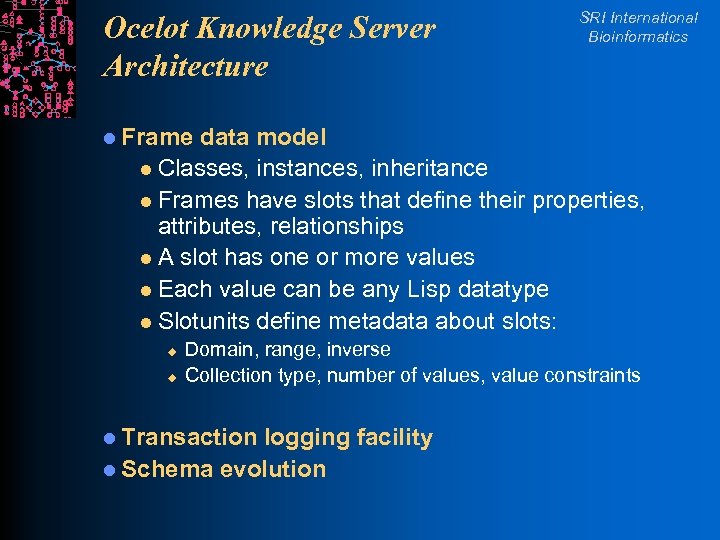
Ocelot Knowledge Server Architecture SRI International Bioinformatics l Frame data model l Classes, instances, inheritance l Frames have slots that define their properties, attributes, relationships l A slot has one or more values l Each value can be any Lisp datatype l Slotunits define metadata about slots: u u Domain, range, inverse Collection type, number of values, value constraints l Transaction logging facility l Schema evolution
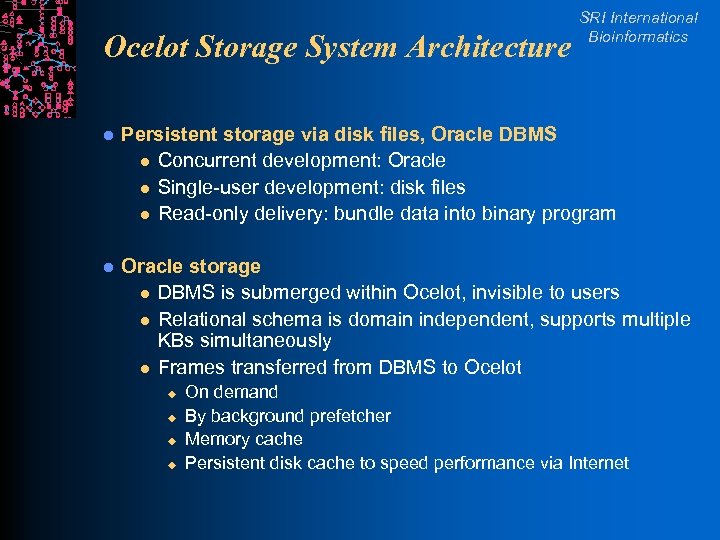
Ocelot Storage System Architecture SRI International Bioinformatics l Persistent storage via disk files, Oracle DBMS l Concurrent development: Oracle l Single-user development: disk files l Read-only delivery: bundle data into binary program l Oracle storage l DBMS is submerged within Ocelot, invisible to users l Relational schema is domain independent, supports multiple KBs simultaneously l Frames transferred from DBMS to Ocelot u u On demand By background prefetcher Memory cache Persistent disk cache to speed performance via Internet
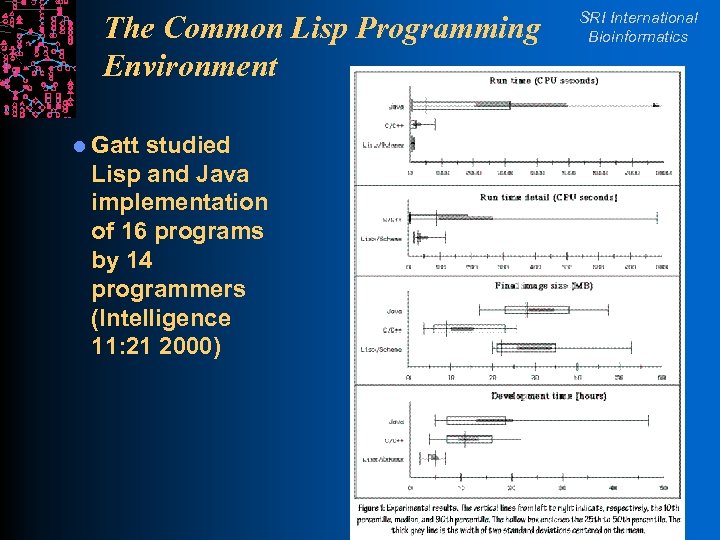
The Common Lisp Programming Environment l Gatt studied Lisp and Java implementation of 16 programs by 14 programmers (Intelligence 11: 21 2000) SRI International Bioinformatics
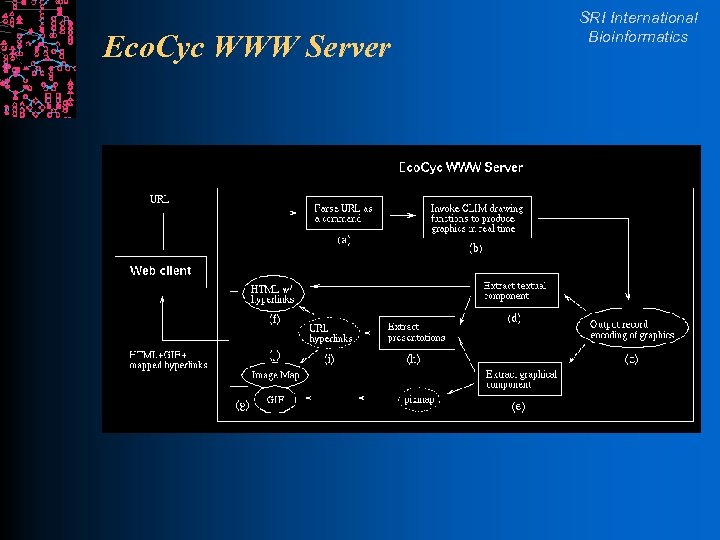
Eco. Cyc WWW Server SRI International Bioinformatics
Pathway/Genome DBs Created by External Users SRI International Bioinformatics l. Plasmodium falciparum, Stanford University l plasmocyc. stanford. edu l. Mycobacterium tuberculosis, Stanford University l Bio. Cyc. org l. Arabidopsis thaliana and Synechosistis, Carnegie Institution of Washington l Arabidopsis. org: 1555 l. Methanococcus l janaschii, EBI Maine. ebi. ac. uk: 1555 l. Other PGDBs in progress by 24 other users l. Software freely available l. Each PGDB owned by its creator

Global Consistency Checking of Biochemical Network SRI International Bioinformatics l Given: A PGDB for an organism l A set of initial metabolites l l Infer: l What set of products can be synthesized by the small-molecule metabolism of the organism l Can known growth medium yield known essential compounds? l Pacific Symposium on Biocomputing p 471 2001
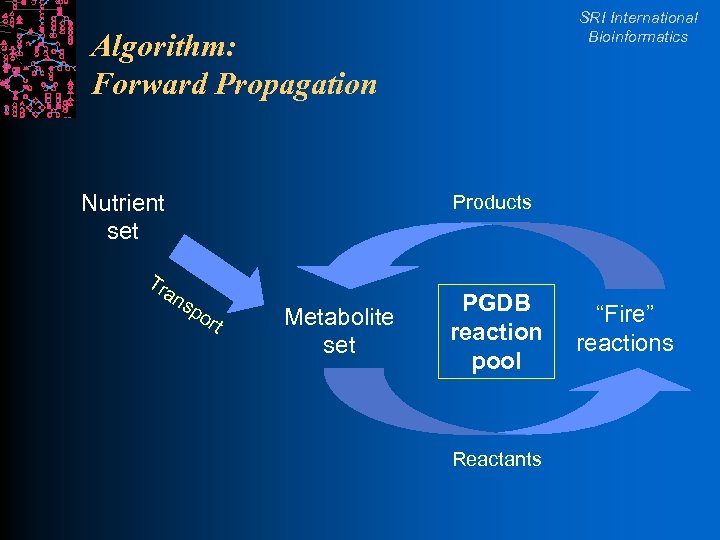
SRI International Bioinformatics Algorithm: Forward Propagation Nutrient set Tr an Products sp o rt Metabolite set PGDB reaction pool Reactants “Fire” reactions

Results SRI International Bioinformatics l Phase I: Forward propagation l 21 initial compounds yielded only half of 38 essential compounds for E. coli l Phase II: Manually identify l Bugs in Eco. Cyc (e. g. , two objects for tryptophan) l Missing initial protein substrates (e. g. , ACP) l Missing pathways in Eco. Cyc l Phase III: Forward propagation with 11 more initial metabolites l Yielded all 38 essential compounds
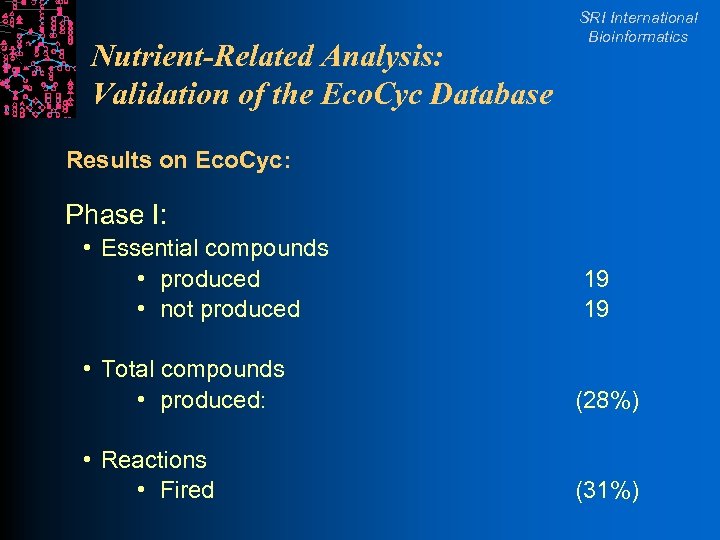
Nutrient-Related Analysis: Validation of the Eco. Cyc Database SRI International Bioinformatics Results on Eco. Cyc: Phase I: • Essential compounds • produced • not produced 19 19 • Total compounds • produced: (28%) • Reactions • Fired (31%)

SRI International Bioinformatics Missing Essential Compounds Due To l Bugs in Eco. Cyc l Narrow conceptualization of the problem l Protein substrates l Incomplete biochemical knowledge

Nutrient-Related Analysis: Validation of the Eco. Cyc Database SRI International Bioinformatics Results on Eco. Cyc: Phase II (After adding 11 extra metabolites): • Essential compounds • produced • not produced • Total compounds • produced: • not produced: • Reactions • Fired • Not fired 38 0 (49%) (51%) (58%) (42%)

Pathway Tools Misconceptions SRI International Bioinformatics l Patho. Logic l Does not re-annotate genomes l Pathway Tools does not handle quantitative information l Pathway/Genome web Editors do not work through the

SRI International Bioinformatics Human. Cyc: Human Metabolic Pathway Database Consortium l Construct DB of human metabolic pathways using Patho. Logic l Link to human genome web sites l Hire one curator to refine and curate with respect to literature over a 2 year period l Remove false-positive predictions l Insert known pathways missed by Patho. Logic l Add comments and citations from pathways and enzymes to the literature l Add enzyme activators, inhibitors, cofactors, tissue information l Available as flatfiles and with Pathway/Genome Navigator

Summary SRI International Bioinformatics l Pathway/Genome Databases l Meta. Cyc non-redundant DB of literature-derived pathways l 14 organism-specific PGDBs available through SRI at Bio. Cyc. org l Computational theories of biochemical machinery l Pathway Tools software l Extract pathways from genomes l Morph annotated genome into structured ontology l Distributed curation tools for MODs l Query, visualization, WWW publishing

Bio. Cyc and Pathway Tools Availability l WWW SRI International Bioinformatics Bio. Cyc freely available to all l Bio. Cyc. org l Six Bio. Cyc DBs openly available to all l Bio. Cyc DBs freely available to non-profits l Flatfiles downloadable from Bio. Cyc. org l Binary executable: u u l Sun Ultra. Sparc-170 w/ 64 MB memory PC, 400 MHz CPU, 64 MB memory, Windows-98 or newer Perl. Cyc API l Pathway Tools freely available to non-profits

SRI International Bioinformatics Acknowledgements l. SRI l Suzanne Paley, Pedro Romero, John Pick, Cindy Krieger, Martha Arnaud l. Eco. Cyc l Project Julio Collado-Vides, Ian Paulsen, Monica Riley, Milton Saier l. Meta. Cyc l Project Sue Rhee, Lukas Mueller, Peifen Zhang, Chris Somerville l. Stanford l Gary Schoolnik, Harley l. Funding sources: l NIH National Center for Research Resources l NIH National Institute of General Medical Sciences l NIH National Human Genome Research Institute l Department of Energy Microbial Cell Project l DARPA Bio. Spice, UPC Bio. Cyc. org
f531b92f1354c0836868dffdefc5cfd3.ppt