076398cb01a64bb47f42848998107ff7.ppt
- Количество слайдов: 96

 The Course of Development
The Course of Development
 The Course of Development Time Events
The Course of Development Time Events
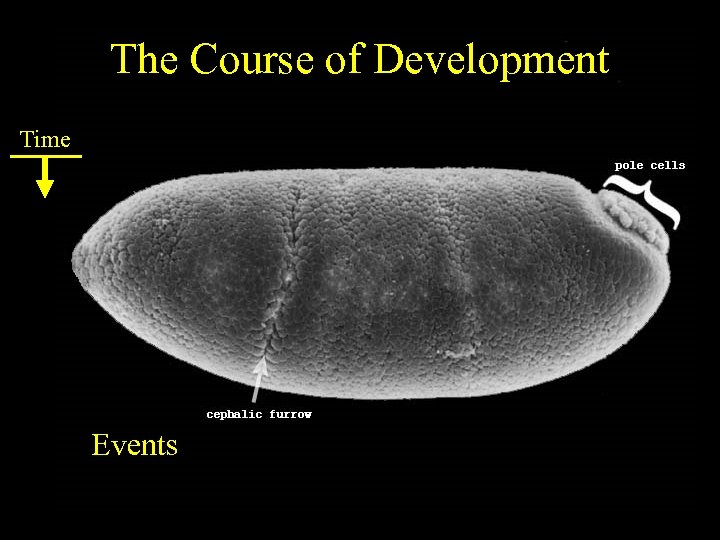 The Course of Development Time Events
The Course of Development Time Events
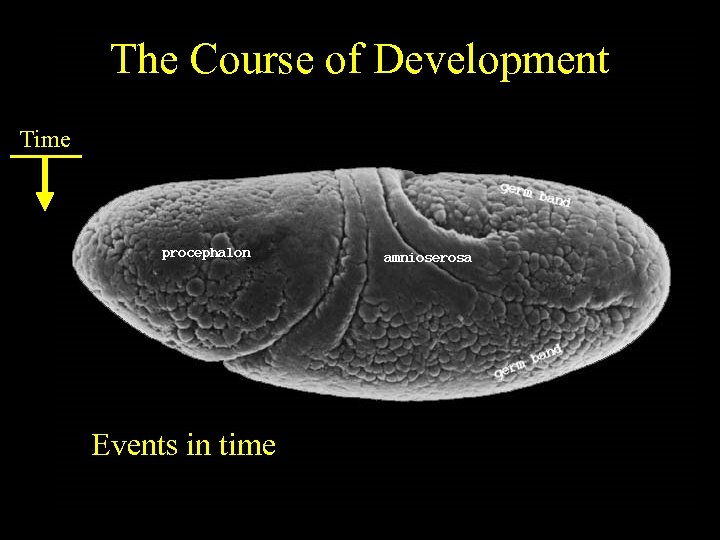 The Course of Development Time Events in time
The Course of Development Time Events in time
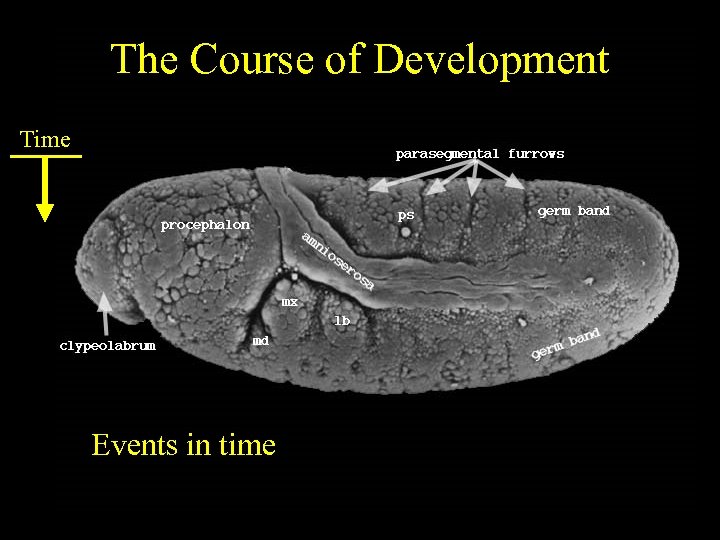 The Course of Development Time Events in time
The Course of Development Time Events in time
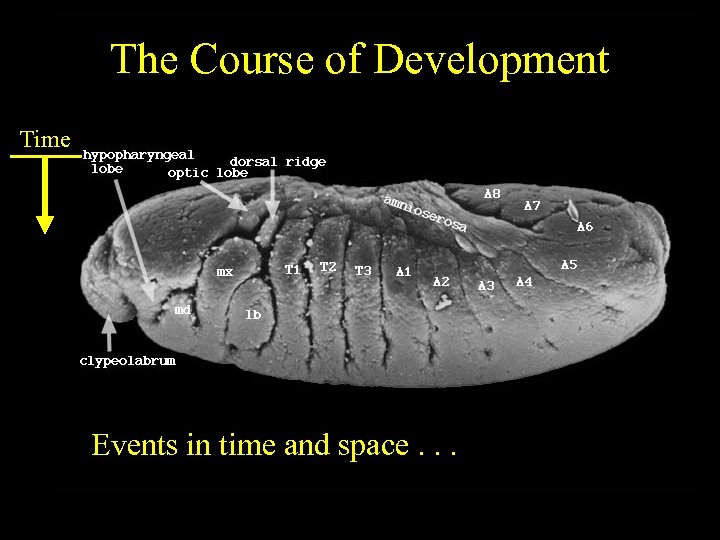 The Course of Development Time Events in time and space. . .
The Course of Development Time Events in time and space. . .
 The Course of Development Time Events in time and space. . .
The Course of Development Time Events in time and space. . .
 The Course of Development Time Events in time and space. . .
The Course of Development Time Events in time and space. . .
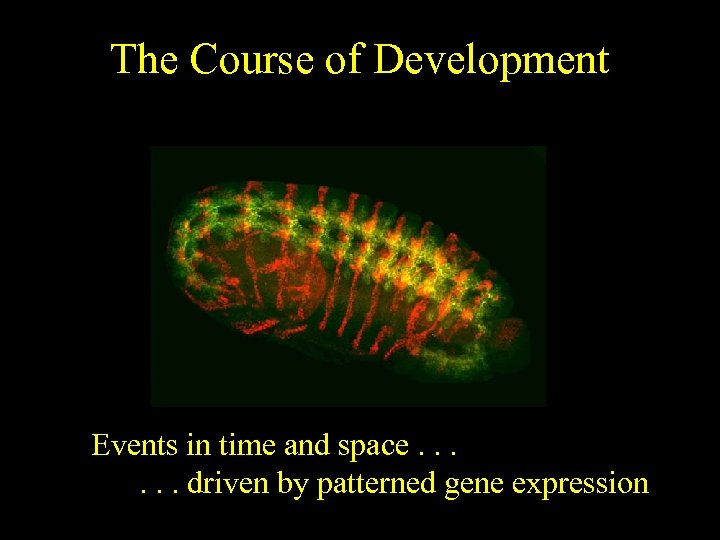 The Course of Development Events in time and space. . . driven by patterned gene expression
The Course of Development Events in time and space. . . driven by patterned gene expression
 The Course of Development Understanding Human Development Events in time and space. . . driven by patterned gene expression
The Course of Development Understanding Human Development Events in time and space. . . driven by patterned gene expression
 The Course of Development Understanding Human Development
The Course of Development Understanding Human Development
 The Course of Development Understanding Human Development
The Course of Development Understanding Human Development
 The Course of Development Understanding Human Development The fate of cells patterned in time and space Intrinsic control? Extrinsic control?
The Course of Development Understanding Human Development The fate of cells patterned in time and space Intrinsic control? Extrinsic control?
 Understanding Human Development Why so difficult? Process 9 mo – 20 yrs Generation 20 yrs Genetic recombination Uncontrolled Genetic manipulation Difficult / Impossible Genome size ~3 billion nucleotides Development Complex How to attack a problem that’s too complex?
Understanding Human Development Why so difficult? Process 9 mo – 20 yrs Generation 20 yrs Genetic recombination Uncontrolled Genetic manipulation Difficult / Impossible Genome size ~3 billion nucleotides Development Complex How to attack a problem that’s too complex?
 How to Attack a Complex Problem Probability of getting a full house?
How to Attack a Complex Problem Probability of getting a full house?
 How to Attack a Complex Problem Probability of getting a pair?
How to Attack a Complex Problem Probability of getting a pair?
 How to Attack a Complex Problem 1 · 3/51 Probability of getting a pair in 2 cards?
How to Attack a Complex Problem 1 · 3/51 Probability of getting a pair in 2 cards?
 Simplification can help in understanding complexity
Simplification can help in understanding complexity
 Understanding Human Development Why so difficult? Process 9 mo – 20 yrs Generation 20 yrs Genetic recombination Uncontrolled Genetic manipulation Difficult / Impossible Genome size ~3 billion nucleotides Development Complex
Understanding Human Development Why so difficult? Process 9 mo – 20 yrs Generation 20 yrs Genetic recombination Uncontrolled Genetic manipulation Difficult / Impossible Genome size ~3 billion nucleotides Development Complex
 Understanding Fly Development Still difficult Process ~8 mo – 20 yrs 9 days Generation 20 days ~14 yrs Genetic recombination Uncontrolled Controlled Genetic manipulation Difficult / Impossible Genome size ~3 billion nucleotides ~170 million nucleotides Development Complex How to simplify further?
Understanding Fly Development Still difficult Process ~8 mo – 20 yrs 9 days Generation 20 days ~14 yrs Genetic recombination Uncontrolled Controlled Genetic manipulation Difficult / Impossible Genome size ~3 billion nucleotides ~170 million nucleotides Development Complex How to simplify further?
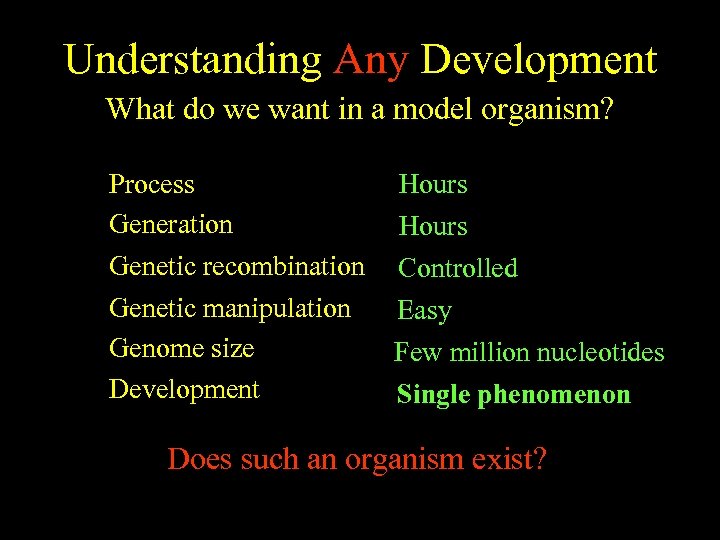 Understanding Any Development What do we want in a model organism? Process ~8 days Hours Generation ~14 days Hours Genetic recombination Controlled Genetic manipulation Difficult Easy Genome size ~170 million nucleotides Few million nucleotides Development Complex Single phenomenon Does such an organism exist?
Understanding Any Development What do we want in a model organism? Process ~8 days Hours Generation ~14 days Hours Genetic recombination Controlled Genetic manipulation Difficult Easy Genome size ~170 million nucleotides Few million nucleotides Development Complex Single phenomenon Does such an organism exist?
 Bacteria . . . but no development
Bacteria . . . but no development
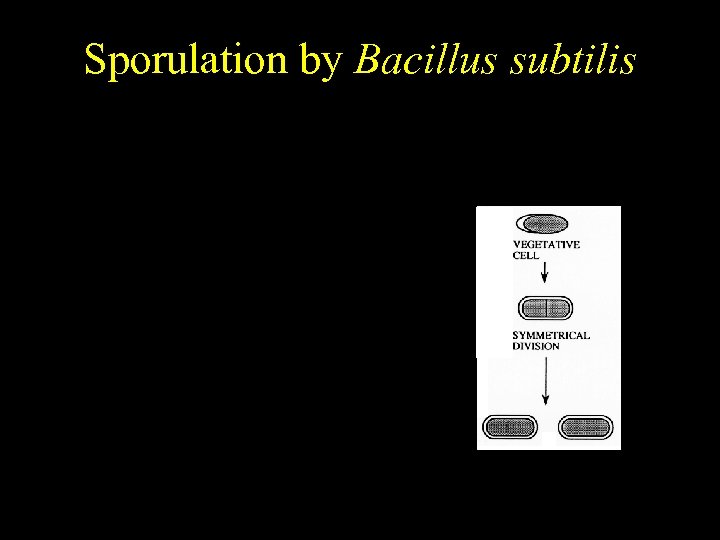 Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation
Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation
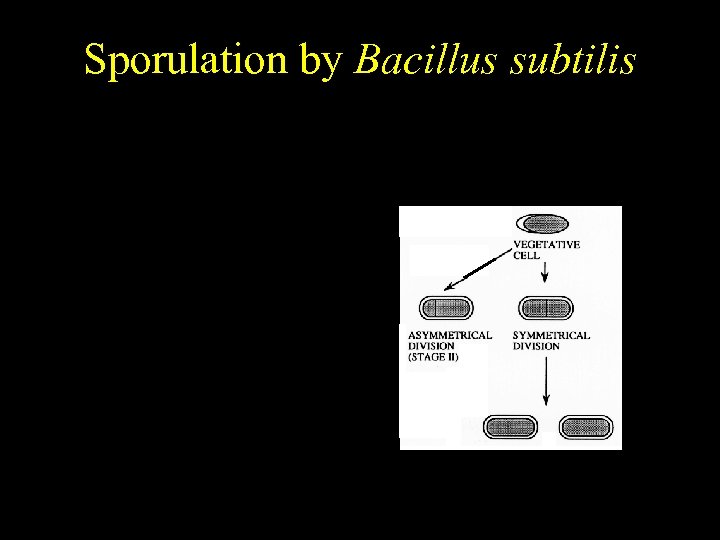 Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation
Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation
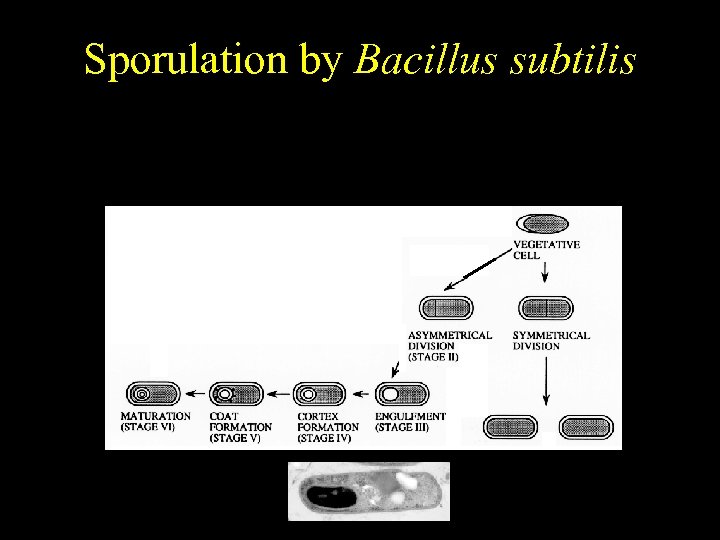 Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation
Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation
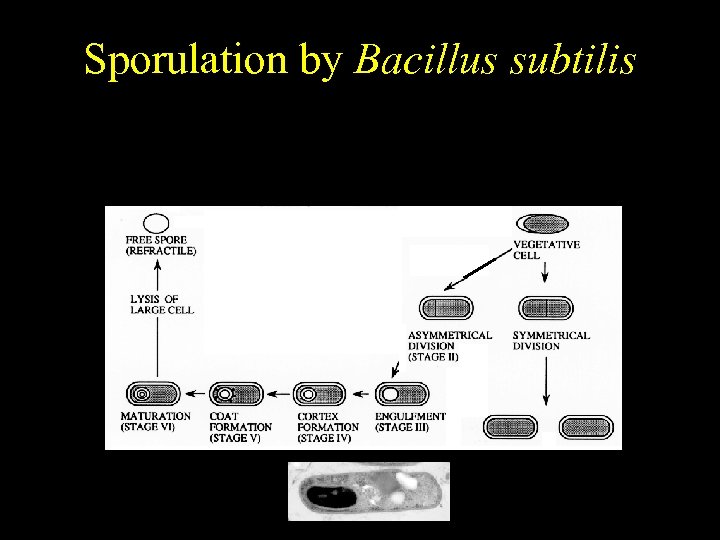 Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation
Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation
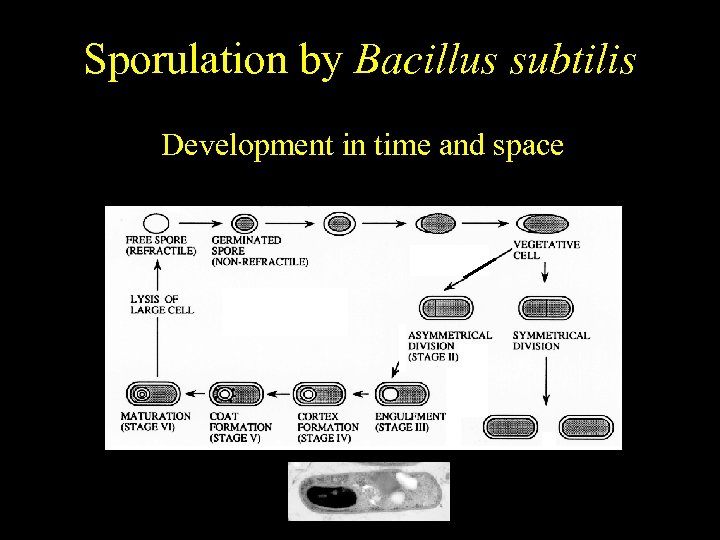 Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation Development in time and space
Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation Development in time and space
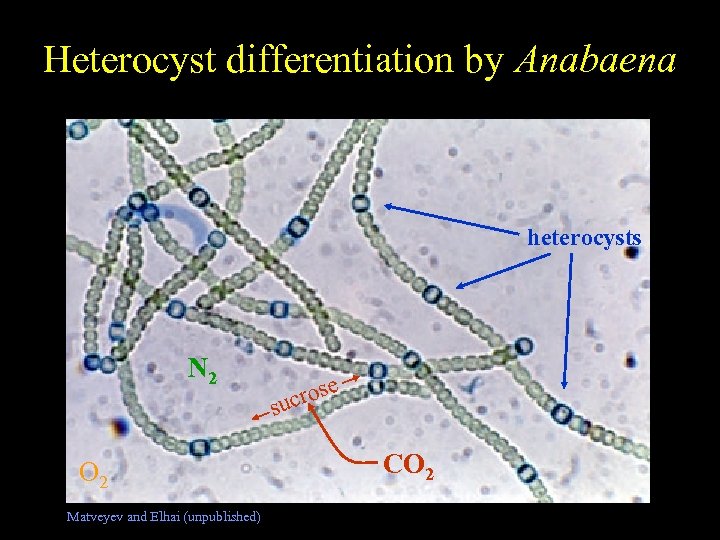 Heterocyst differentiation by Anabaena Free-living Nostoc heterocysts N 2 O 2 Matveyev and Elhai (unpublished) rose suc CO 2
Heterocyst differentiation by Anabaena Free-living Nostoc heterocysts N 2 O 2 Matveyev and Elhai (unpublished) rose suc CO 2
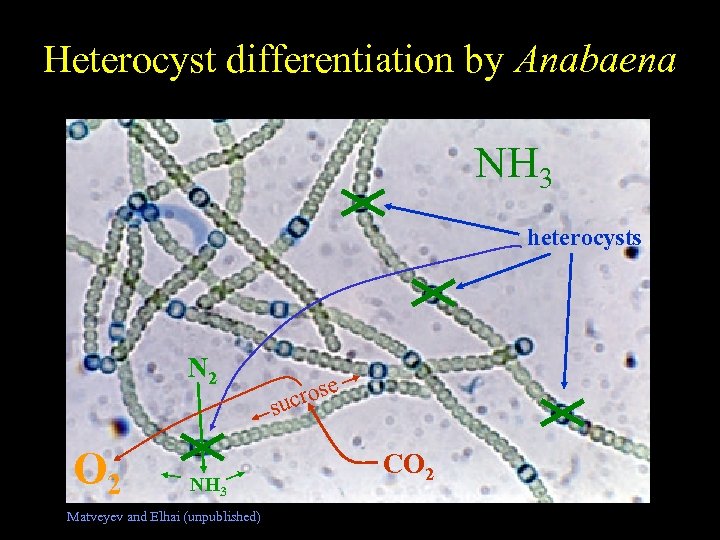 Heterocyst differentiation by Anabaena Free-living Nostoc NH 3 heterocysts N 2 O 2 NH 3 Matveyev and Elhai (unpublished) rose suc CO 2
Heterocyst differentiation by Anabaena Free-living Nostoc NH 3 heterocysts N 2 O 2 NH 3 Matveyev and Elhai (unpublished) rose suc CO 2
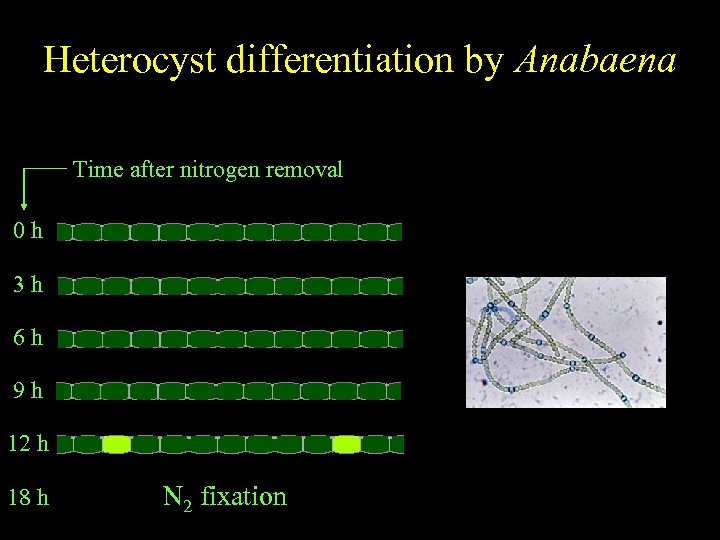 Anabaena Heterocyst differentiation by Anabaena Spatially regulated differentiation Time after nitrogen removal 0 h 3 h 6 h 9 h 12 h 18 h N 2 fixation
Anabaena Heterocyst differentiation by Anabaena Spatially regulated differentiation Time after nitrogen removal 0 h 3 h 6 h 9 h 12 h 18 h N 2 fixation
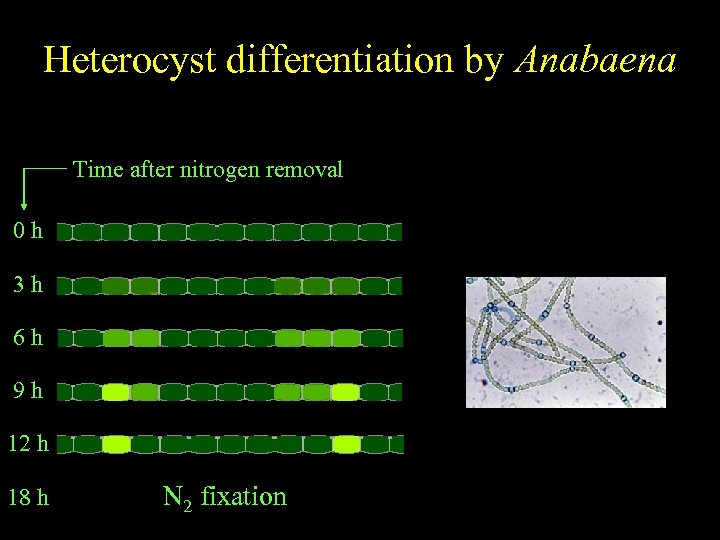 Anabaena Heterocyst differentiation by Anabaena Spatially regulated differentiation Time after nitrogen removal 0 h 3 h 6 h 9 h 12 h 18 h N 2 fixation
Anabaena Heterocyst differentiation by Anabaena Spatially regulated differentiation Time after nitrogen removal 0 h 3 h 6 h 9 h 12 h 18 h N 2 fixation
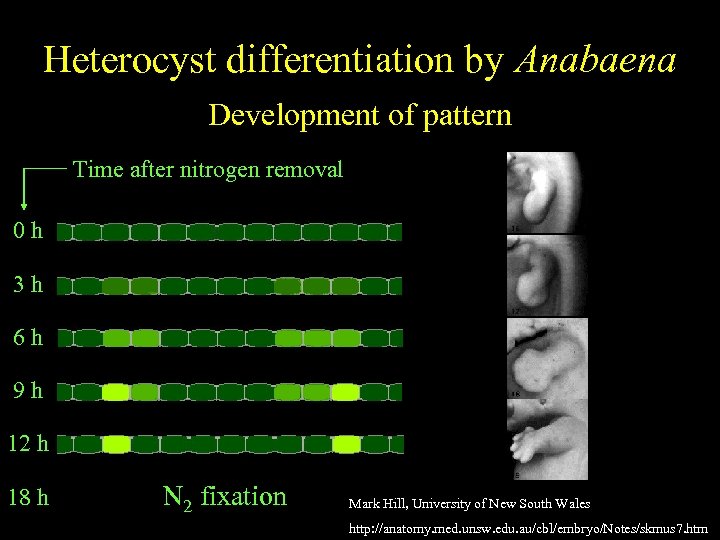 Anabaena Heterocyst differentiation by Anabaena Spatially regulatedof pattern differentiation Development Time after nitrogen removal 0 h 3 h 6 h 9 h 12 h 18 h N 2 fixation Mark Hill, University of New South Wales http: //anatomy. med. unsw. edu. au/cbl/embryo/Notes/skmus 7. htm
Anabaena Heterocyst differentiation by Anabaena Spatially regulatedof pattern differentiation Development Time after nitrogen removal 0 h 3 h 6 h 9 h 12 h 18 h N 2 fixation Mark Hill, University of New South Wales http: //anatomy. med. unsw. edu. au/cbl/embryo/Notes/skmus 7. htm
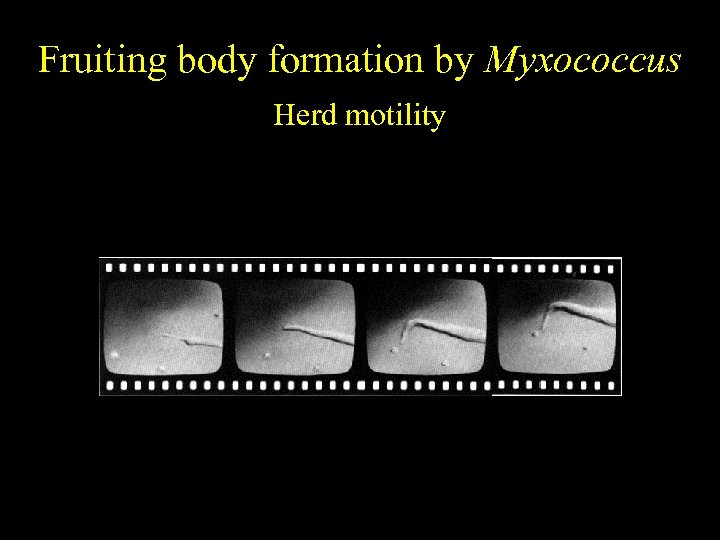 Fruiting body formation by Myxococcus Herd motility
Fruiting body formation by Myxococcus Herd motility
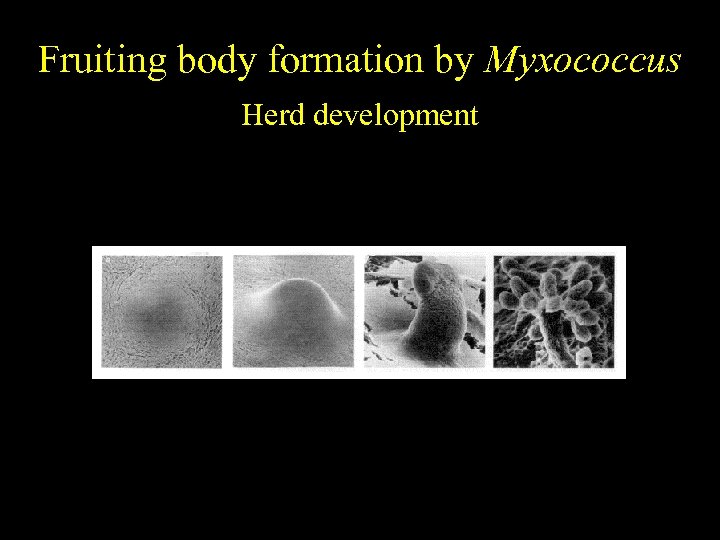 Fruiting body formation by Myxococcus Herd development
Fruiting body formation by Myxococcus Herd development
 Fruiting body formation by Myxococcus Extrinsic control over development
Fruiting body formation by Myxococcus Extrinsic control over development
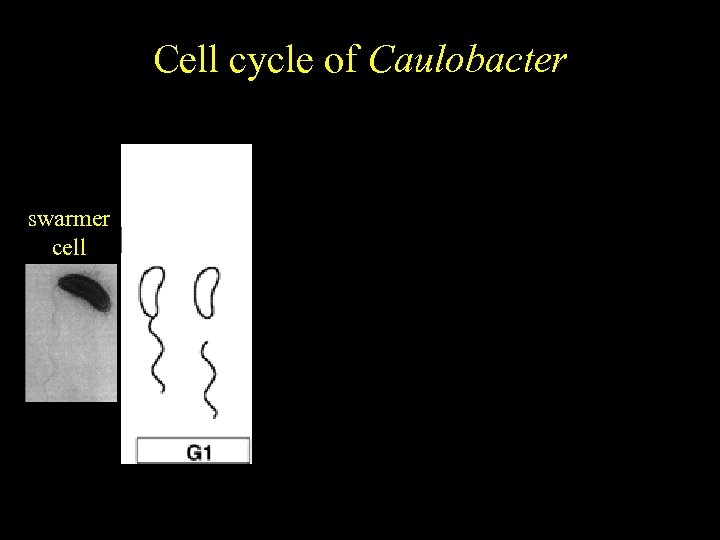 Caulobacter Cell cycle of crescentus Cell cycle-regulated differentiation swarmer cell
Caulobacter Cell cycle of crescentus Cell cycle-regulated differentiation swarmer cell
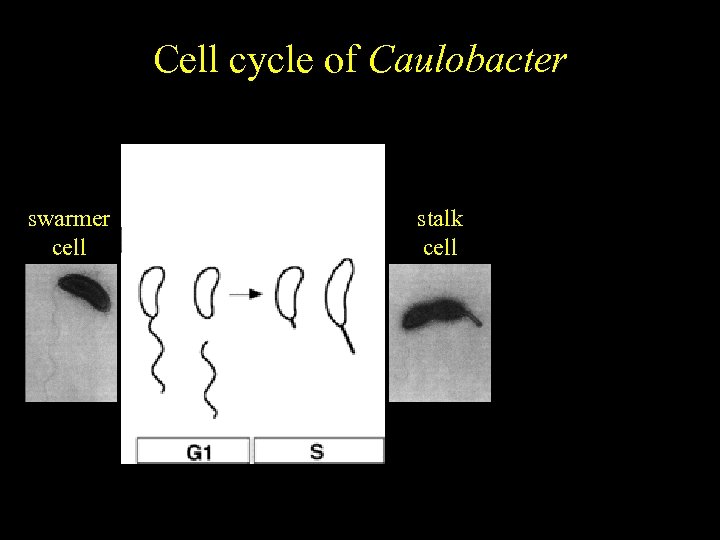 Caulobacter Cell cycle of crescentus Cell cycle-regulated differentiation swarmer cell stalk cell
Caulobacter Cell cycle of crescentus Cell cycle-regulated differentiation swarmer cell stalk cell
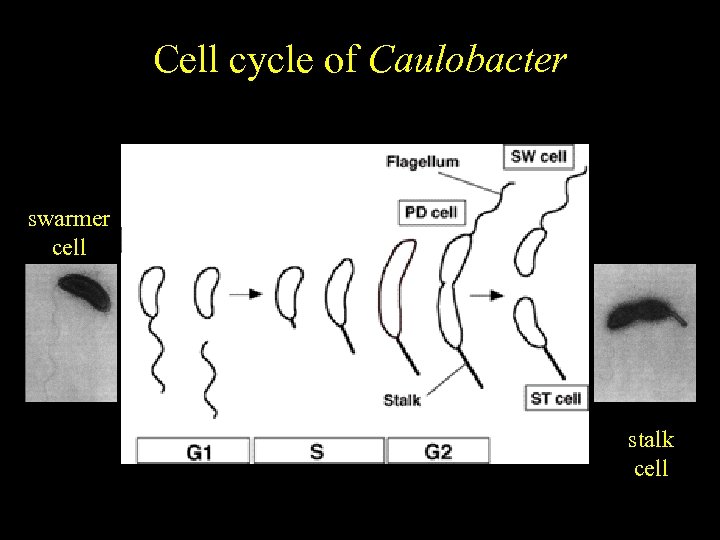 Caulobacter Cell cycle of crescentus Cell cycle-regulated differentiation swarmer cell stalk cell
Caulobacter Cell cycle of crescentus Cell cycle-regulated differentiation swarmer cell stalk cell
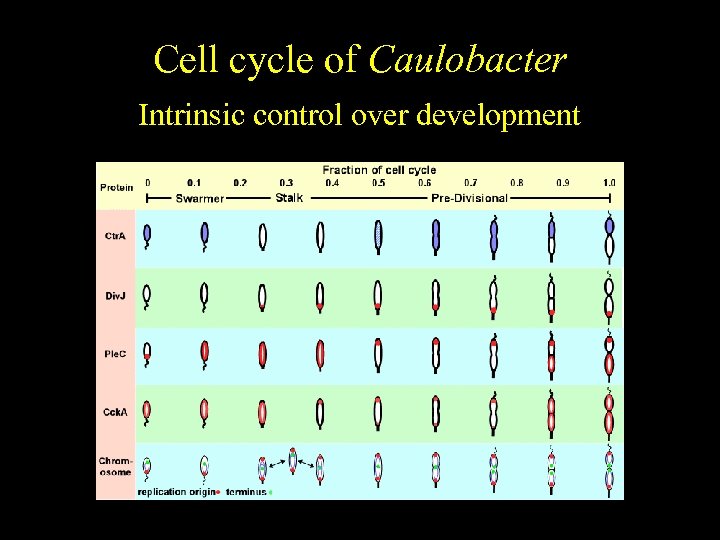 Caulobacter Cell cycle of crescentus Cell cycle-regulated differentiation Intrinsic control over development
Caulobacter Cell cycle of crescentus Cell cycle-regulated differentiation Intrinsic control over development
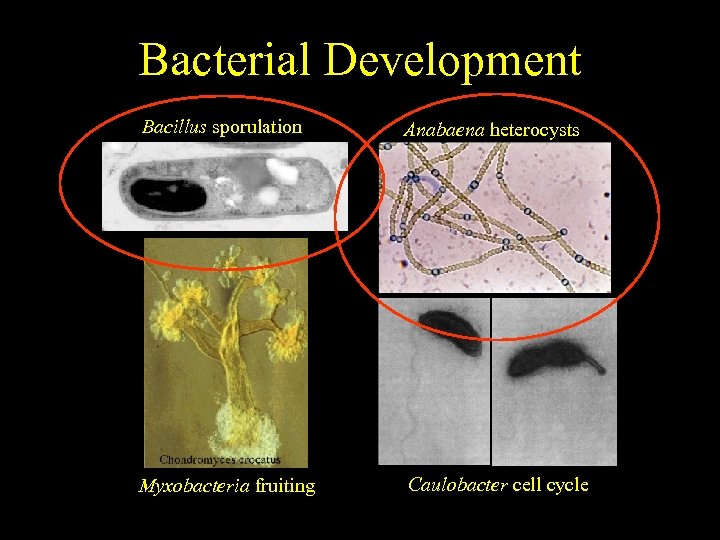 Bacterial Development End result. . . much simpler Bacillus sporulation Anabaena heterocysts Myxobacteria fruiting Caulobacter cell cycle
Bacterial Development End result. . . much simpler Bacillus sporulation Anabaena heterocysts Myxobacteria fruiting Caulobacter cell cycle
 Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation Control of initiation selective gene expression How to make the decision? ?
Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation Control of initiation selective gene expression How to make the decision? ?
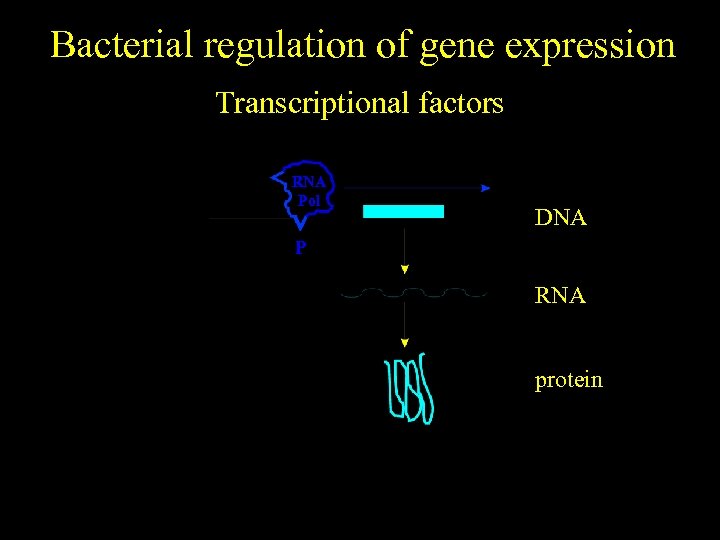 Bacterial regulation of gene expression Transcriptional factors RNA Pol DNA P RNA protein
Bacterial regulation of gene expression Transcriptional factors RNA Pol DNA P RNA protein
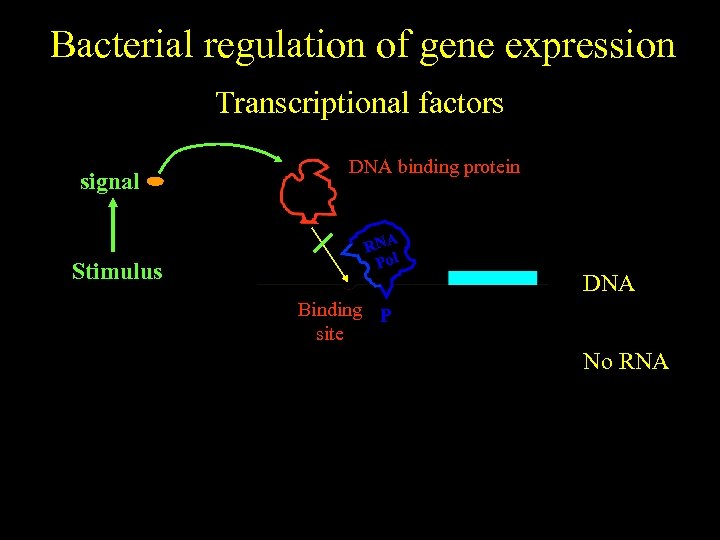 Bacterial regulation of gene expression Transcriptional factors signal No stimulus Stimulus DNA binding protein RNA Pol DNA Binding P site No RNA
Bacterial regulation of gene expression Transcriptional factors signal No stimulus Stimulus DNA binding protein RNA Pol DNA Binding P site No RNA
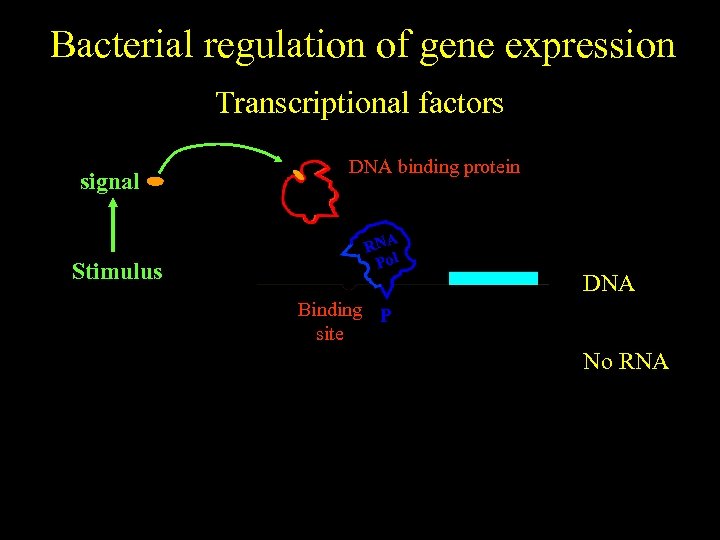 Bacterial regulation of gene expression Transcriptional factors signal No stimulus Stimulus DNA binding protein RNA Pol DNA Binding P site No RNA
Bacterial regulation of gene expression Transcriptional factors signal No stimulus Stimulus DNA binding protein RNA Pol DNA Binding P site No RNA
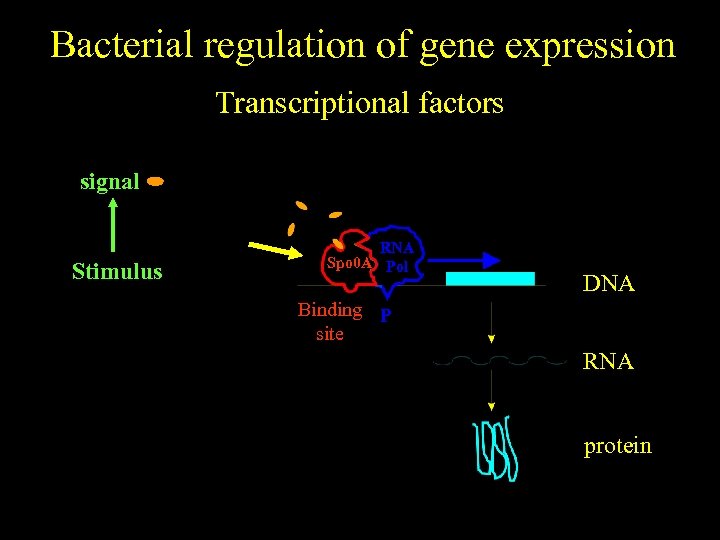 Bacterial regulation of gene expression Transcriptional factors signal No stimulus Stimulus RNA Spo 0 A Pol DNA Binding P site RNA protein
Bacterial regulation of gene expression Transcriptional factors signal No stimulus Stimulus RNA Spo 0 A Pol DNA Binding P site RNA protein
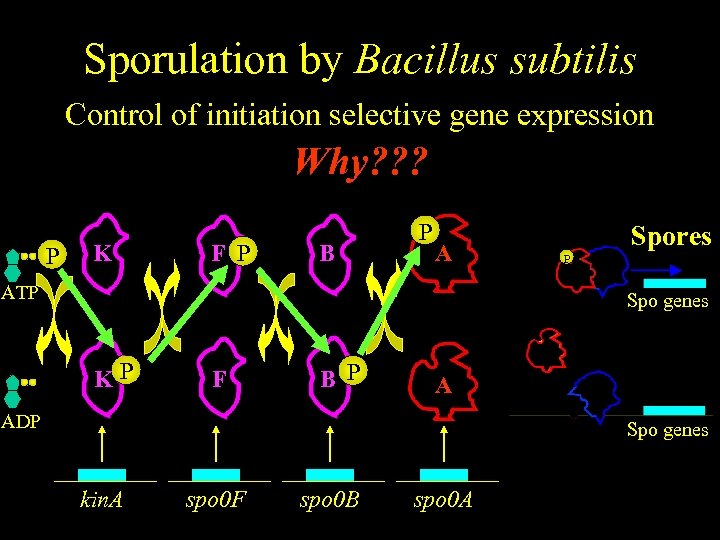 Sporulation by Bacillus subtilis Control of initiation selective gene expression Why? ? ? P K F P B P A ATP Spores P Spo genes KP F B P A ADP Spo genes kin. A spo 0 F spo 0 B spo 0 A
Sporulation by Bacillus subtilis Control of initiation selective gene expression Why? ? ? P K F P B P A ATP Spores P Spo genes KP F B P A ADP Spo genes kin. A spo 0 F spo 0 B spo 0 A
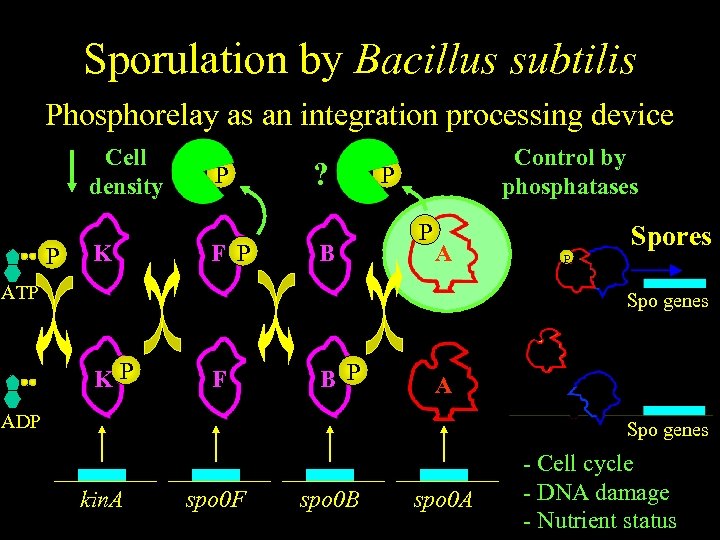 Sporulation by Bacillus subtilis Phosphorelay as an integration processing device Cell density P K P F P ? B Control by phosphatases P P A ATP Spores P Spo genes KP F B P A ADP Spo genes kin. A spo 0 F spo 0 B spo 0 A - Cell cycle - DNA damage - Nutrient status
Sporulation by Bacillus subtilis Phosphorelay as an integration processing device Cell density P K P F P ? B Control by phosphatases P P A ATP Spores P Spo genes KP F B P A ADP Spo genes kin. A spo 0 F spo 0 B spo 0 A - Cell cycle - DNA damage - Nutrient status
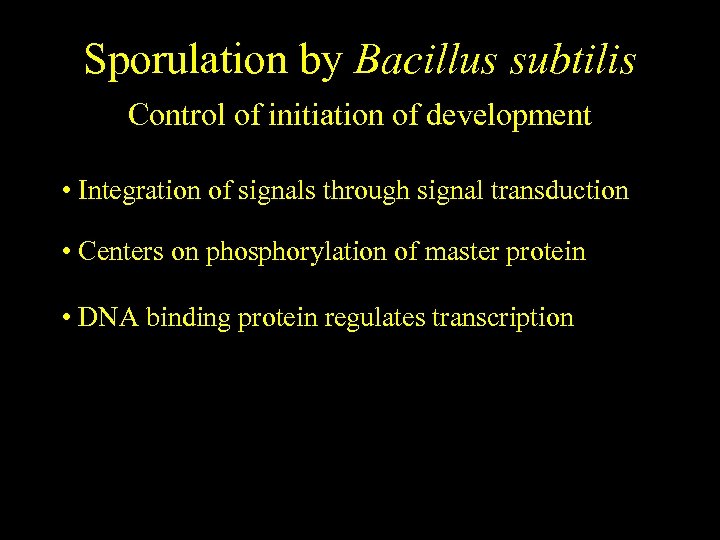 Sporulation by Bacillus subtilis Control of initiation of development • Integration of signals through signal transduction • Centers on phosphorylation of master protein • DNA binding protein regulates transcription
Sporulation by Bacillus subtilis Control of initiation of development • Integration of signals through signal transduction • Centers on phosphorylation of master protein • DNA binding protein regulates transcription
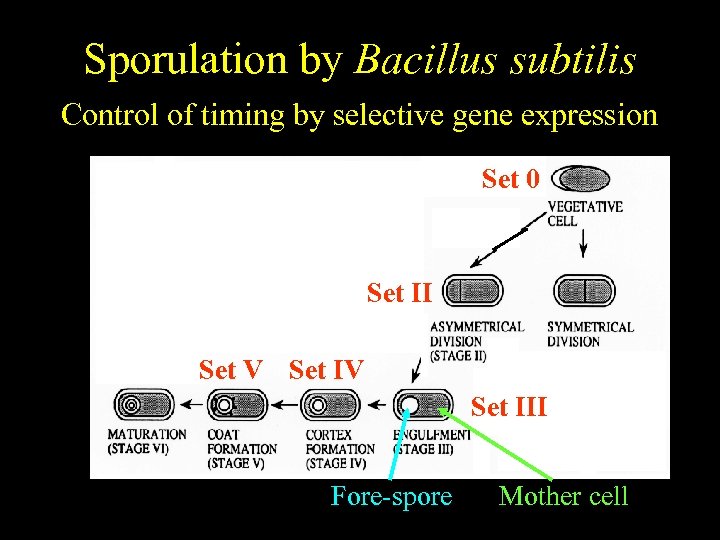 Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation Control of timing by selective gene expression Set 0 Set II Set V Set III Fore-spore Mother cell
Bacillus subtilis Sporulation by Bacillus subtilis Temporally regulated differentiation Control of timing by selective gene expression Set 0 Set II Set V Set III Fore-spore Mother cell
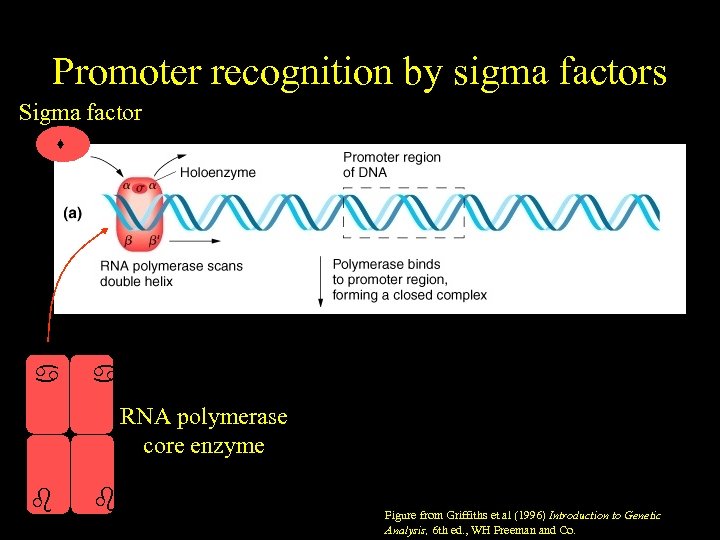 Promoter recognition by sigma factors Sigma factor RNA polymerase core enzyme ' Figure from Griffiths et al (1996) Introduction to Genetic Analysis, 6 th ed. , WH Freeman and Co.
Promoter recognition by sigma factors Sigma factor RNA polymerase core enzyme ' Figure from Griffiths et al (1996) Introduction to Genetic Analysis, 6 th ed. , WH Freeman and Co.
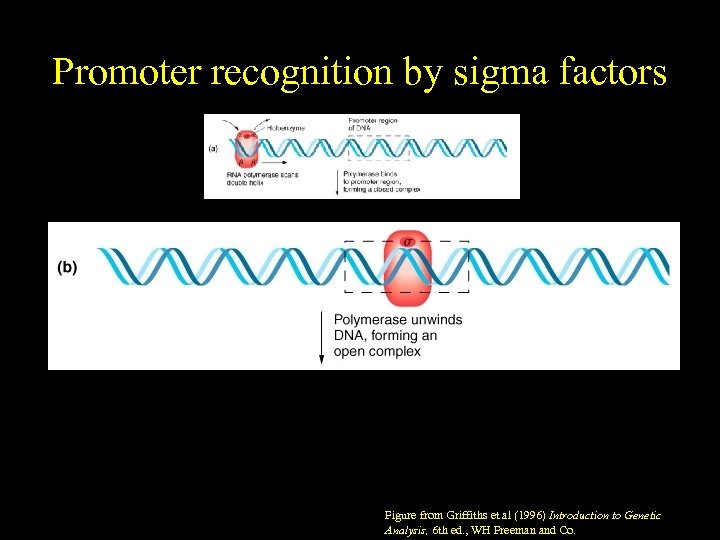 Promoter recognition by sigma factors Figure from Griffiths et al (1996) Introduction to Genetic Analysis, 6 th ed. , WH Freeman and Co.
Promoter recognition by sigma factors Figure from Griffiths et al (1996) Introduction to Genetic Analysis, 6 th ed. , WH Freeman and Co.
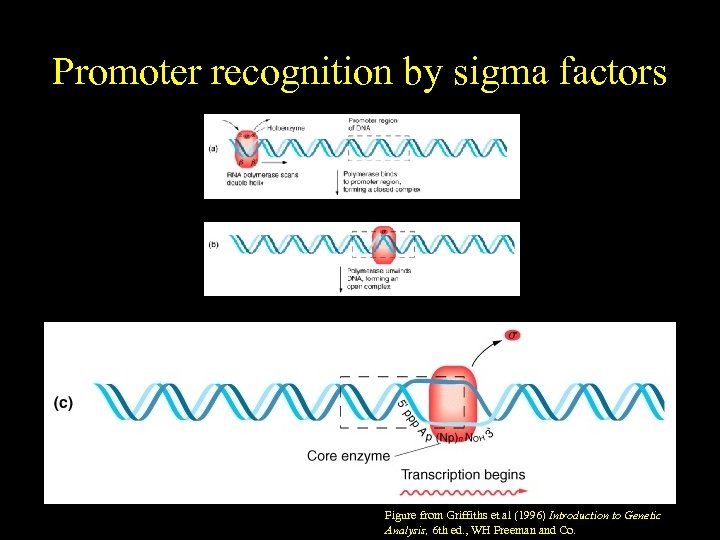 Promoter recognition by sigma factors Figure from Griffiths et al (1996) Introduction to Genetic Analysis, 6 th ed. , WH Freeman and Co.
Promoter recognition by sigma factors Figure from Griffiths et al (1996) Introduction to Genetic Analysis, 6 th ed. , WH Freeman and Co.
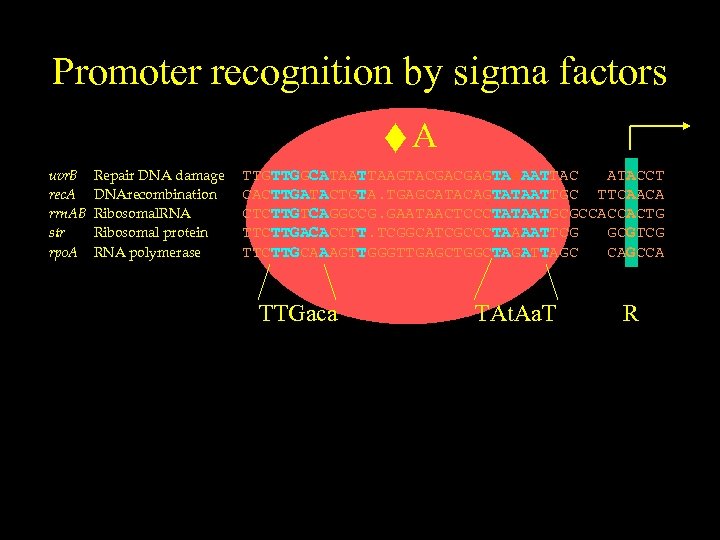 Promoter recognition by sigma factors uvr. B rec. A rrn. AB str rpo. A Repair DNA damage DNArecombination Ribosomal. RNA Ribosomal protein RNA polymerase A TTGTTGGCATAATTAAGTACGACGAGTAAAATTAC ATACCT CACTTGATACTGTA. TGAGCATACAGTATAATTGC TTCAACA CTCTTGTCAGGCCG. GAATAACTCCCTATAATGCGCCACCACTG TTCTTGACACCTT. TCGGCATCGCCCTAAAATTCG GCGTCG TTCTTGCAAAGTTGGGTTGAGCTGGCTAGATTAGC CAGCCA TTGaca TAt. Aa. T R
Promoter recognition by sigma factors uvr. B rec. A rrn. AB str rpo. A Repair DNA damage DNArecombination Ribosomal. RNA Ribosomal protein RNA polymerase A TTGTTGGCATAATTAAGTACGACGAGTAAAATTAC ATACCT CACTTGATACTGTA. TGAGCATACAGTATAATTGC TTCAACA CTCTTGTCAGGCCG. GAATAACTCCCTATAATGCGCCACCACTG TTCTTGACACCTT. TCGGCATCGCCCTAAAATTCG GCGTCG TTCTTGCAAAGTTGGGTTGAGCTGGCTAGATTAGC CAGCCA TTGaca TAt. Aa. T R
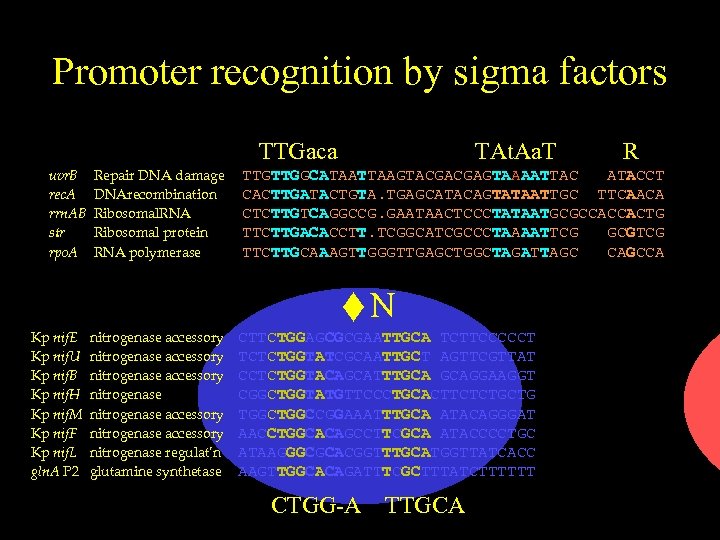 Promoter recognition by sigma factors TTGaca uvr. B rec. A rrn. AB str rpo. A Kp nif. E Kp nif. U Kp nif. B Kp nif. H Kp nif. M Kp nif. F Kp nif. L gln. A P 2 Repair DNA damage DNArecombination Ribosomal. RNA Ribosomal protein RNA polymerase nitrogenase accessory nitrogenase accessory nitrogenase regulat’n glutamine synthetase TAt. Aa. T R TTGTTGGCATAATTAAGTACGACGAGTAAAATTAC ATACCT CACTTGATACTGTA. TGAGCATACAGTATAATTGC TTCAACA CTCTTGTCAGGCCG. GAATAACTCCCTATAATGCGCCACCACTG TTCTTGACACCTT. TCGGCATCGCCCTAAAATTCG GCGTCG TTCTTGCAAAGTTGGGTTGAGCTGGCTAGATTAGC CAGCCA N CTTCTGGAGCGCGAATTGCA TCTTCCCCCT TCTCTGGTATCGCAATTGCT AGTTCGTTAT CCTCTGGTACAGCATTTGCA GCAGGAAGGT CGGCTGGTATGTTCCCTGCACTTCTCTGCTG TGGCCGGAAATTTGCA ATACAGGGAT AACCTGGCACAGCCTTCGCA ATACCCCTGC ATAAGGGCGCACGGTTTGCATGGTTATCACC AAGTTGGCACAGATTTCGCTTTATCTTTTTT CTGG-A TTGCA
Promoter recognition by sigma factors TTGaca uvr. B rec. A rrn. AB str rpo. A Kp nif. E Kp nif. U Kp nif. B Kp nif. H Kp nif. M Kp nif. F Kp nif. L gln. A P 2 Repair DNA damage DNArecombination Ribosomal. RNA Ribosomal protein RNA polymerase nitrogenase accessory nitrogenase accessory nitrogenase regulat’n glutamine synthetase TAt. Aa. T R TTGTTGGCATAATTAAGTACGACGAGTAAAATTAC ATACCT CACTTGATACTGTA. TGAGCATACAGTATAATTGC TTCAACA CTCTTGTCAGGCCG. GAATAACTCCCTATAATGCGCCACCACTG TTCTTGACACCTT. TCGGCATCGCCCTAAAATTCG GCGTCG TTCTTGCAAAGTTGGGTTGAGCTGGCTAGATTAGC CAGCCA N CTTCTGGAGCGCGAATTGCA TCTTCCCCCT TCTCTGGTATCGCAATTGCT AGTTCGTTAT CCTCTGGTACAGCATTTGCA GCAGGAAGGT CGGCTGGTATGTTCCCTGCACTTCTCTGCTG TGGCCGGAAATTTGCA ATACAGGGAT AACCTGGCACAGCCTTCGCA ATACCCCTGC ATAAGGGCGCACGGTTTGCATGGTTATCACC AAGTTGGCACAGATTTCGCTTTATCTTTTTT CTGG-A TTGCA
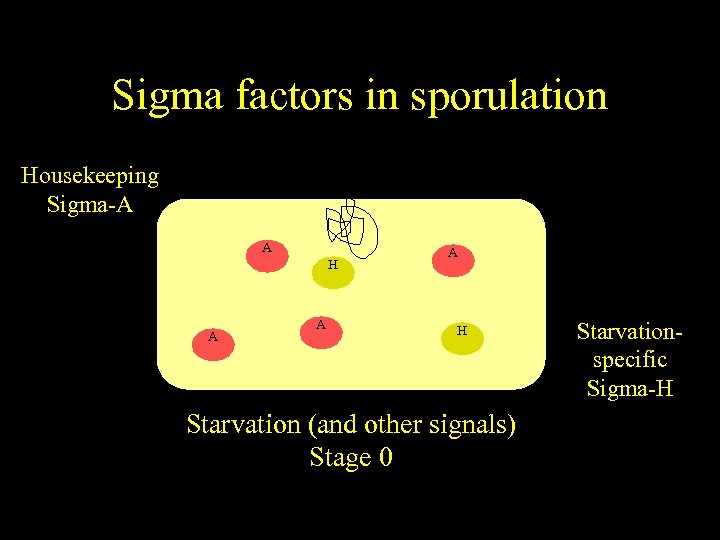 Sigma factors in sporulation Housekeeping Sigma-A A H A A A H Starvation (and other signals) Stage 0 Starvationspecific Sigma-H
Sigma factors in sporulation Housekeeping Sigma-A A H A A A H Starvation (and other signals) Stage 0 Starvationspecific Sigma-H
 Sigma factors in sporulation Mother-specific Mother cell Sigma-E E Forespore A H F A E A A Stage II/III H F Forespecific Sigma-F
Sigma factors in sporulation Mother-specific Mother cell Sigma-E E Forespore A H F A E A A Stage II/III H F Forespecific Sigma-F
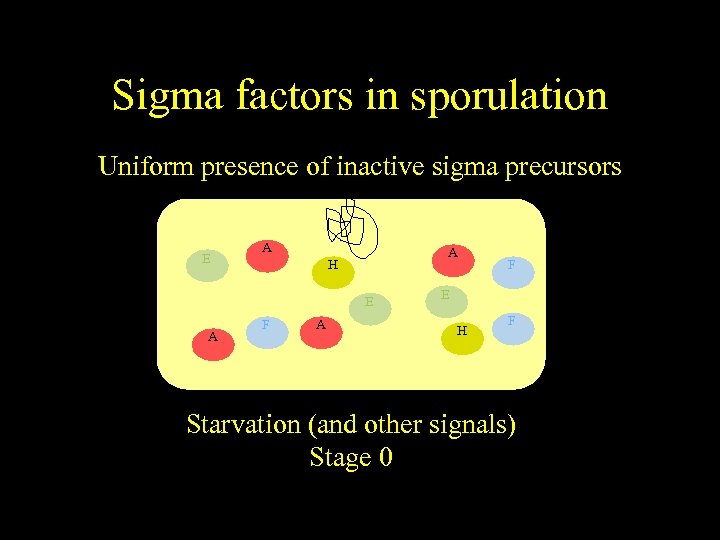 Sigma factors in sporulation Uniform presence of inactive sigma precursors E A A H E A F E H F Starvation (and other signals) Stage 0
Sigma factors in sporulation Uniform presence of inactive sigma precursors E A A H E A F E H F Starvation (and other signals) Stage 0
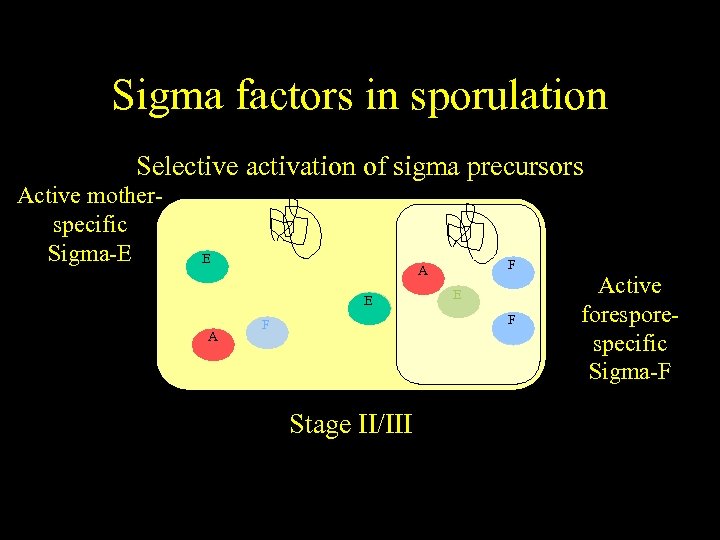 Sigma factors in sporulation Selective activation of sigma precursors Active motherspecific Sigma-E E A H E A F F A A Stage II/III E H F Active forespecific Sigma-F
Sigma factors in sporulation Selective activation of sigma precursors Active motherspecific Sigma-E E A H E A F F A A Stage II/III E H F Active forespecific Sigma-F
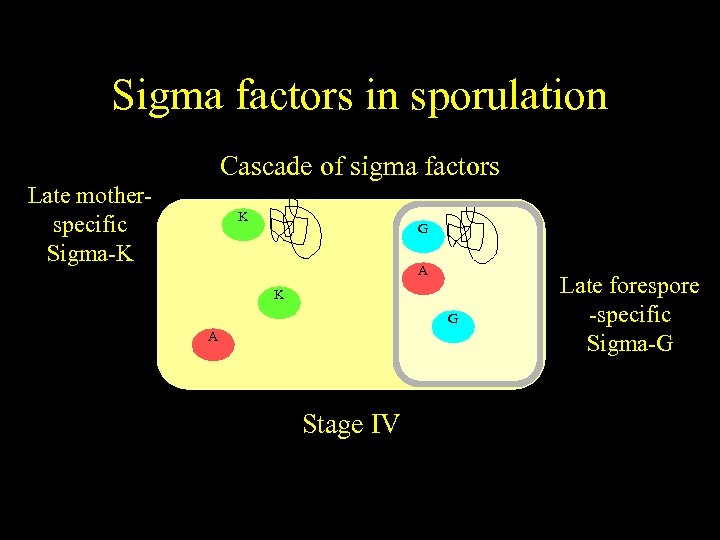 Sigma factors in sporulation Cascade of sigma factors Late motherspecific Sigma-K K G E F A K E G F A Starvation Stageother signals) (and IV Stage III Late forespore -specific Sigma-G
Sigma factors in sporulation Cascade of sigma factors Late motherspecific Sigma-K K G E F A K E G F A Starvation Stageother signals) (and IV Stage III Late forespore -specific Sigma-G
 Sporulation by Bacillus subtilis Control of timing by selective gene expression • Determined by specific, active sigma factors • Presence and activation important • Activation linked to morphological events
Sporulation by Bacillus subtilis Control of timing by selective gene expression • Determined by specific, active sigma factors • Presence and activation important • Activation linked to morphological events
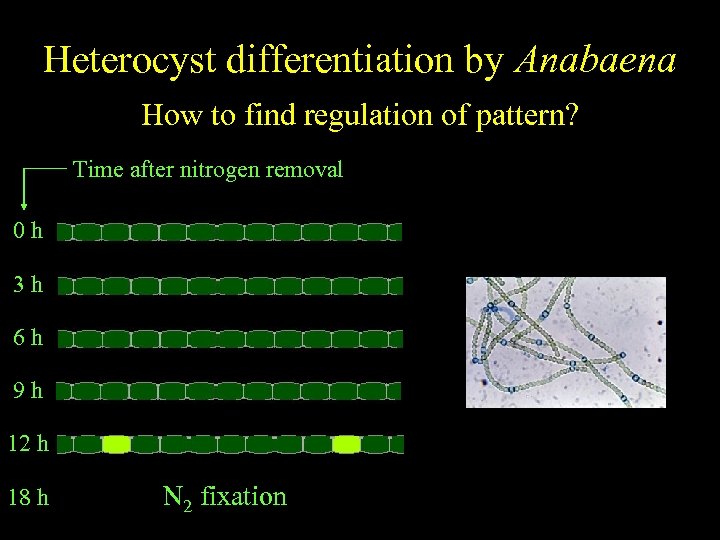 Anabaena Heterocyst differentiation by Anabaena Spatiallyfind regulation of pattern? regulated differentiation How to Time after nitrogen removal 0 h 3 h 6 h 9 h 12 h 18 h N 2 fixation
Anabaena Heterocyst differentiation by Anabaena Spatiallyfind regulation of pattern? regulated differentiation How to Time after nitrogen removal 0 h 3 h 6 h 9 h 12 h 18 h N 2 fixation
 Genetic approach to Cell Biology
Genetic approach to Cell Biology
 Genetic approach to Cell Biology
Genetic approach to Cell Biology
 Genetic approach to Cell Biology
Genetic approach to Cell Biology
 Genetic approach to Cell Biology
Genetic approach to Cell Biology
 Genetic approach to Cell Biology
Genetic approach to Cell Biology
 Genetic approach to Cell Biology
Genetic approach to Cell Biology
 Genetic approach to Cell Biology
Genetic approach to Cell Biology
 Genetic approach to Cell Biology
Genetic approach to Cell Biology
 Genetic approach to Cell Biology Isolation of Defective Gene
Genetic approach to Cell Biology Isolation of Defective Gene
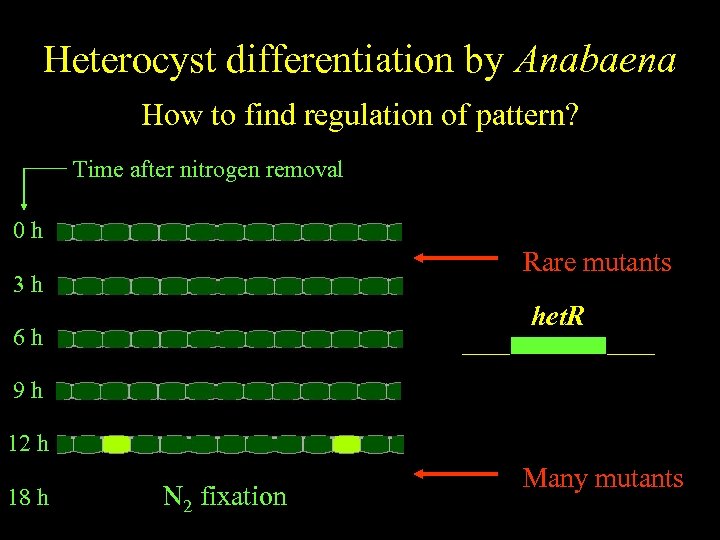 Anabaena Heterocyst differentiation by Anabaena Spatiallyfind regulation of pattern? regulated differentiation How to Time after nitrogen removal 0 h Rare mutants 3 h het. R 6 h 9 h 12 h 18 h N 2 fixation Many mutants
Anabaena Heterocyst differentiation by Anabaena Spatiallyfind regulation of pattern? regulated differentiation How to Time after nitrogen removal 0 h Rare mutants 3 h het. R 6 h 9 h 12 h 18 h N 2 fixation Many mutants
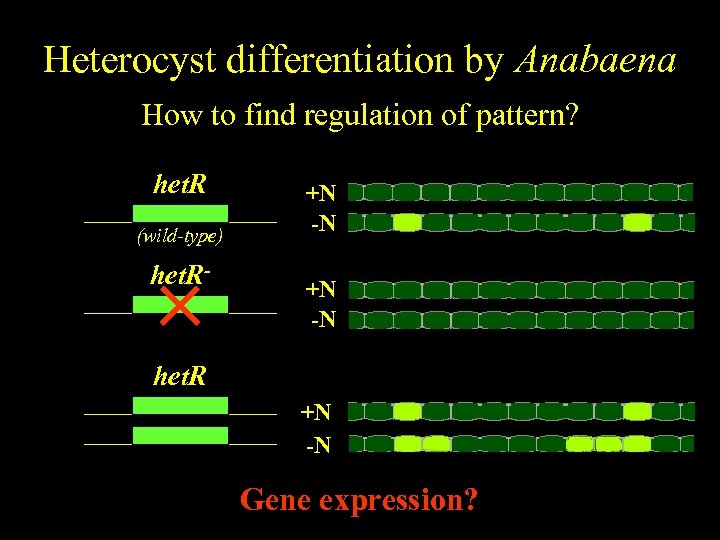 Anabaena Heterocyst differentiation by Anabaena Spatiallyfind regulation of pattern? regulated differentiation How to het. R (wild-type) het. R- +N -N het. R +N -N Gene expression?
Anabaena Heterocyst differentiation by Anabaena Spatiallyfind regulation of pattern? regulated differentiation How to het. R (wild-type) het. R- +N -N het. R +N -N Gene expression?
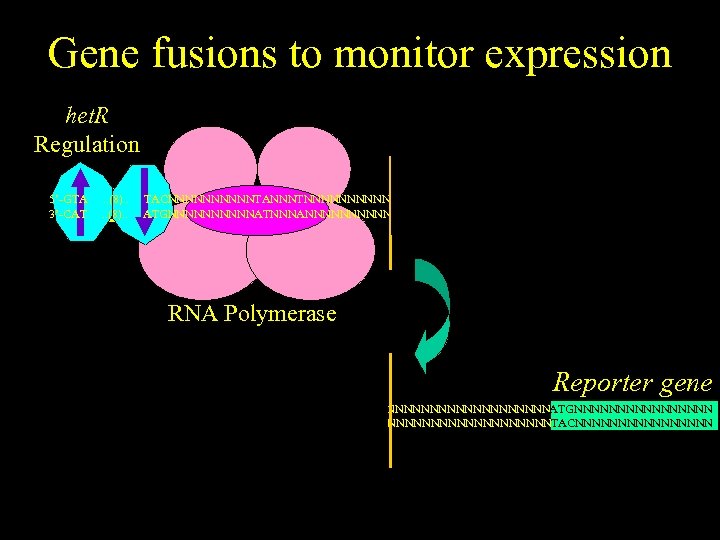 Gene fusions to monitor expression het. R Regulation het. R gene 5’-GTA 3’-CAT . . (8). . TACNNNNNTANNNTNNNNNNNNNNNNNNATGNNNNNNNN ATGNNNNNATNNNANNNNNNNNNNNNNNTACNNNNNNNN RNA Polymerase Reporter gene 5’-GTGAGTTAGCTCACNNNNNTANNNTNNNNNNNNNNNNNNATGNNNNNNNN 3’-CACTCAATCGAGTGNNNNNATNNNANNNNNNNNNNNNNNNTACNNNNNNNN GTA . . (8). . TAC
Gene fusions to monitor expression het. R Regulation het. R gene 5’-GTA 3’-CAT . . (8). . TACNNNNNTANNNTNNNNNNNNNNNNNNATGNNNNNNNN ATGNNNNNATNNNANNNNNNNNNNNNNNTACNNNNNNNN RNA Polymerase Reporter gene 5’-GTGAGTTAGCTCACNNNNNTANNNTNNNNNNNNNNNNNNATGNNNNNNNN 3’-CACTCAATCGAGTGNNNNNATNNNANNNNNNNNNNNNNNNTACNNNNNNNN GTA . . (8). . TAC
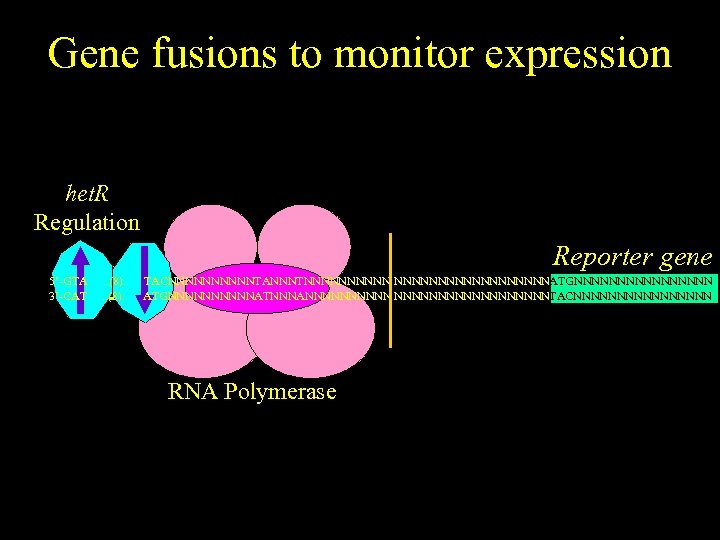 Gene fusions to monitor expression het. R Regulation Reporter gene 5’-GTA 3’-CAT . . (8). . TACNNNNNTANNNTNNNNNNNNNATGNNNNNNNN ATGNNNNNATNNNANNNNNNNNNTACNNNNNNNN RNA Polymerase GTA . . (8). . TAC
Gene fusions to monitor expression het. R Regulation Reporter gene 5’-GTA 3’-CAT . . (8). . TACNNNNNTANNNTNNNNNNNNNATGNNNNNNNN ATGNNNNNATNNNANNNNNNNNNTACNNNNNNNN RNA Polymerase GTA . . (8). . TAC
 Detection of het. R gene expression through Green Fluorescent Protein The hydromedusa Aequoria victoria Source of Green Fluorescent Protein
Detection of het. R gene expression through Green Fluorescent Protein The hydromedusa Aequoria victoria Source of Green Fluorescent Protein
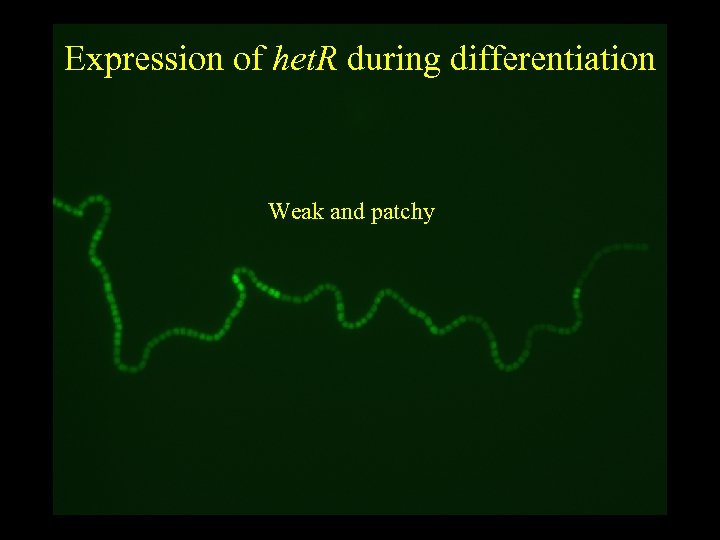 Expression of het. R during differentiation Weak and patchy
Expression of het. R during differentiation Weak and patchy
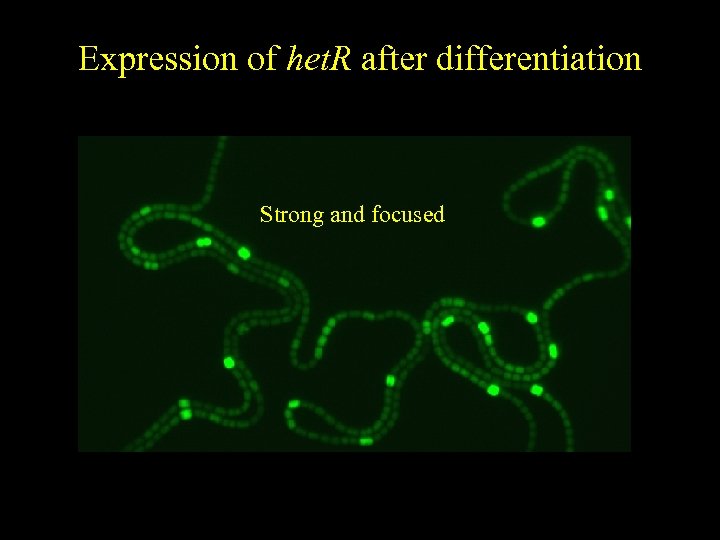 Expression of het. R after differentiation Strong and focused
Expression of het. R after differentiation Strong and focused
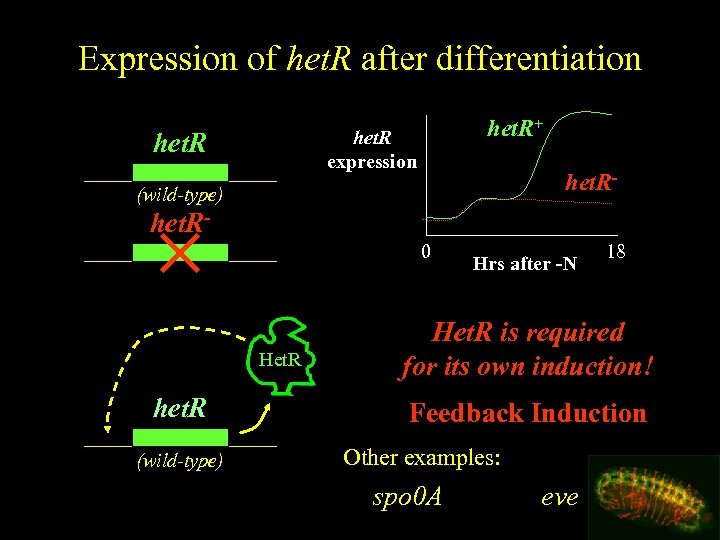 Expression of het. R after differentiation het. R+ het. R expression het. R- (wild-type) het. R 0 Het. R het. R (wild-type) Hrs after -N 18 Het. R is required for its own induction! Feedback Induction Other examples: spo 0 A eve
Expression of het. R after differentiation het. R+ het. R expression het. R- (wild-type) het. R 0 Het. R het. R (wild-type) Hrs after -N 18 Het. R is required for its own induction! Feedback Induction Other examples: spo 0 A eve
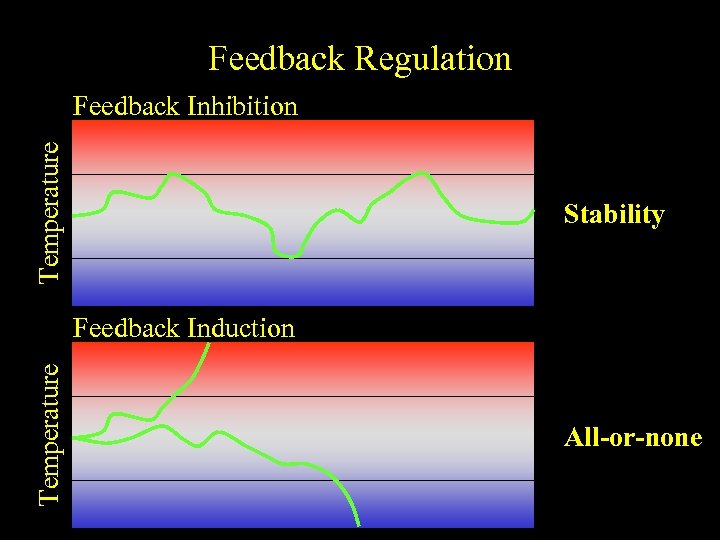 Feedback Regulation Temperature Feedback Inhibition Stability Temperature Feedback Induction All-or-none
Feedback Regulation Temperature Feedback Inhibition Stability Temperature Feedback Induction All-or-none
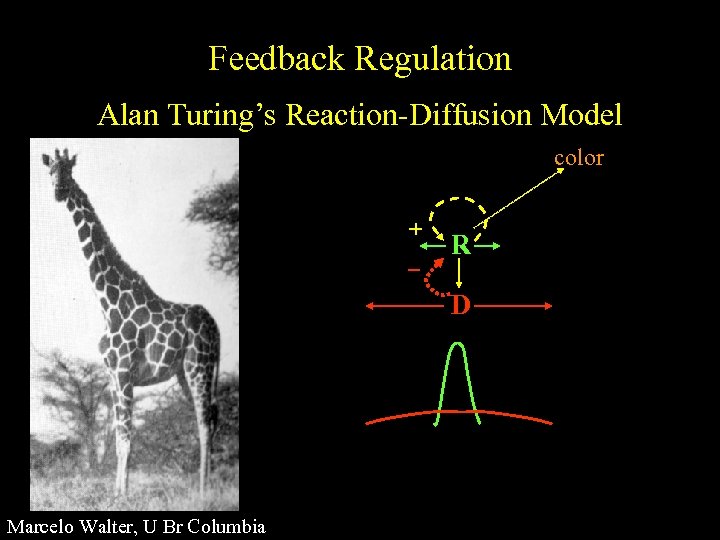 Feedback Regulation Alan Turing’s Reaction-Diffusion Model color + R D Marcelo Walter, U Br Columbia
Feedback Regulation Alan Turing’s Reaction-Diffusion Model color + R D Marcelo Walter, U Br Columbia
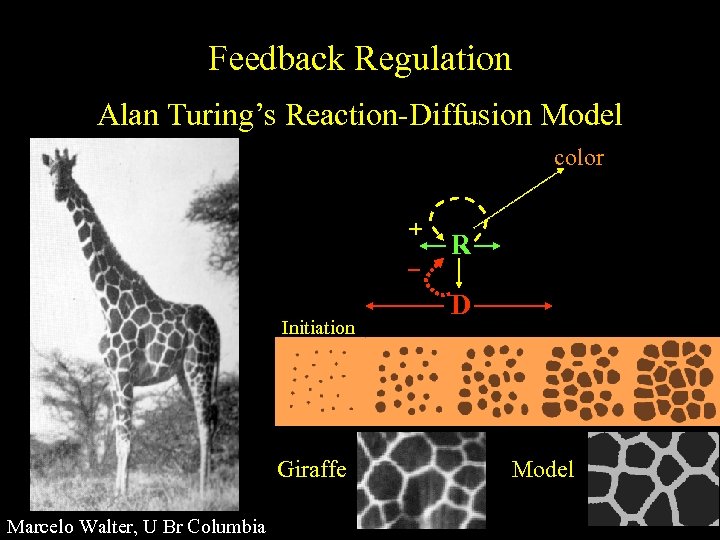 Feedback Regulation Alan Turing’s Reaction-Diffusion Model color + Initiation Giraffe Marcelo Walter, U Br Columbia R D Model
Feedback Regulation Alan Turing’s Reaction-Diffusion Model color + Initiation Giraffe Marcelo Walter, U Br Columbia R D Model
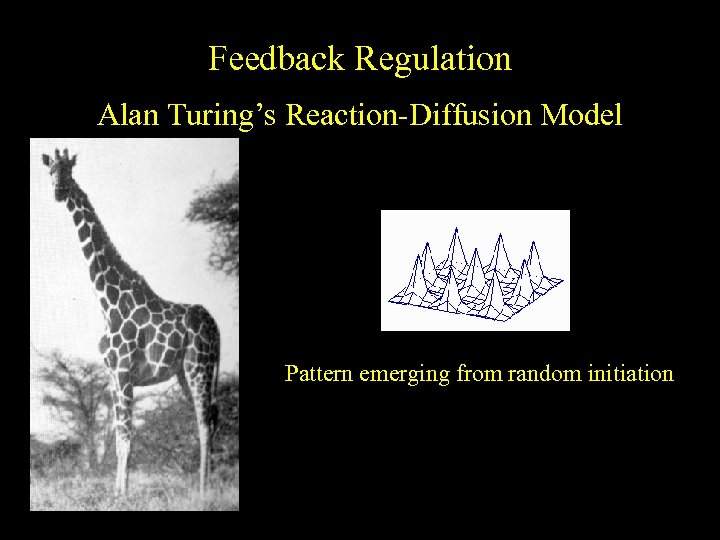 Feedback Regulation Alan Turing’s Reaction-Diffusion Model Pattern emerging from random initiation
Feedback Regulation Alan Turing’s Reaction-Diffusion Model Pattern emerging from random initiation
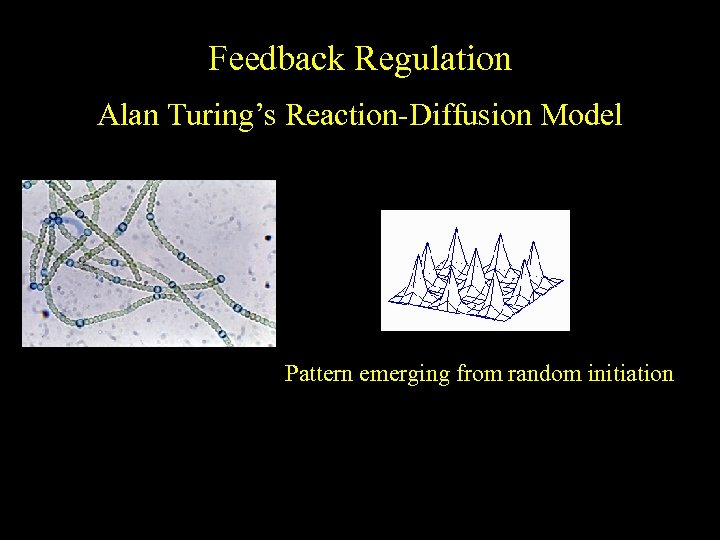 Feedback Regulation Alan Turing’s Reaction-Diffusion Model Pattern emerging from random initiation
Feedback Regulation Alan Turing’s Reaction-Diffusion Model Pattern emerging from random initiation
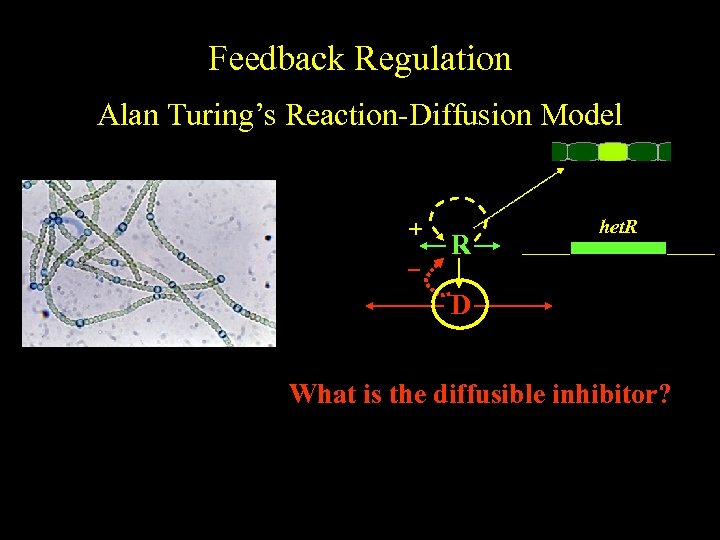 Feedback Regulation Alan Turing’s Reaction-Diffusion Model color + R het. R D What is the diffusible inhibitor?
Feedback Regulation Alan Turing’s Reaction-Diffusion Model color + R het. R D What is the diffusible inhibitor?
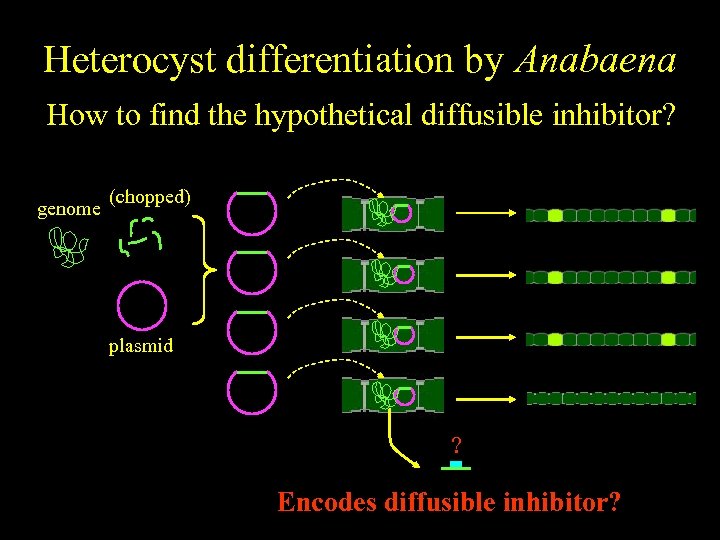 Heterocyst differentiation by Anabaena How to find the hypothetical diffusible inhibitor? genome (chopped) plasmid ? Encodes diffusible inhibitor?
Heterocyst differentiation by Anabaena How to find the hypothetical diffusible inhibitor? genome (chopped) plasmid ? Encodes diffusible inhibitor?
 Heterocyst differentiation by Anabaena The nature of the hypothetical inhibitor (typical size of gene) Pat. S Active part of sequence MLVNFCDERGSGR Is Pat. S the predicted diffusible inhibitor?
Heterocyst differentiation by Anabaena The nature of the hypothetical inhibitor (typical size of gene) Pat. S Active part of sequence MLVNFCDERGSGR Is Pat. S the predicted diffusible inhibitor?
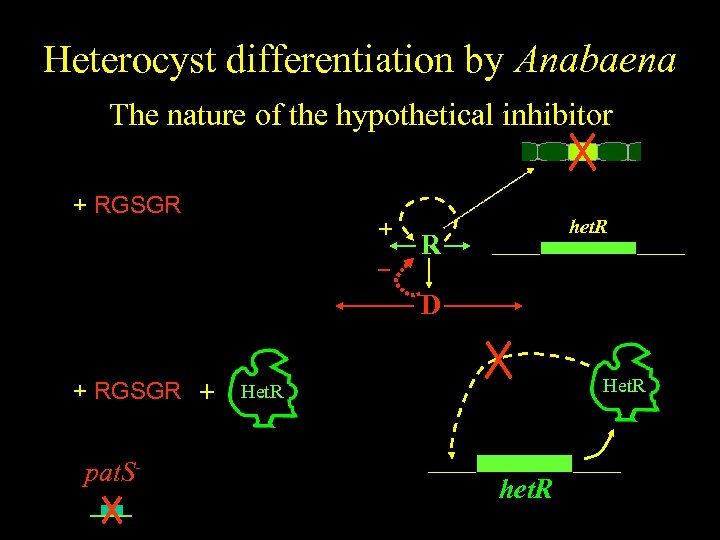 Heterocyst differentiation by Anabaena The nature of the hypothetical inhibitor color + RGSGR + het. R R D + RGSGR + pat. S- Het. R het. R
Heterocyst differentiation by Anabaena The nature of the hypothetical inhibitor color + RGSGR + het. R R D + RGSGR + pat. S- Het. R het. R
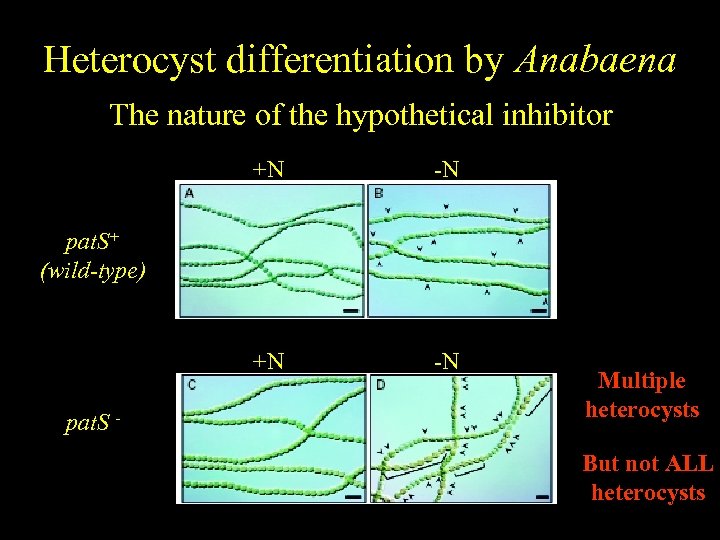 Heterocyst differentiation by Anabaena The nature of the hypothetical inhibitor +N -N pat. S+ (wild-type) pat. S - Multiple heterocysts But not ALL heterocysts
Heterocyst differentiation by Anabaena The nature of the hypothetical inhibitor +N -N pat. S+ (wild-type) pat. S - Multiple heterocysts But not ALL heterocysts
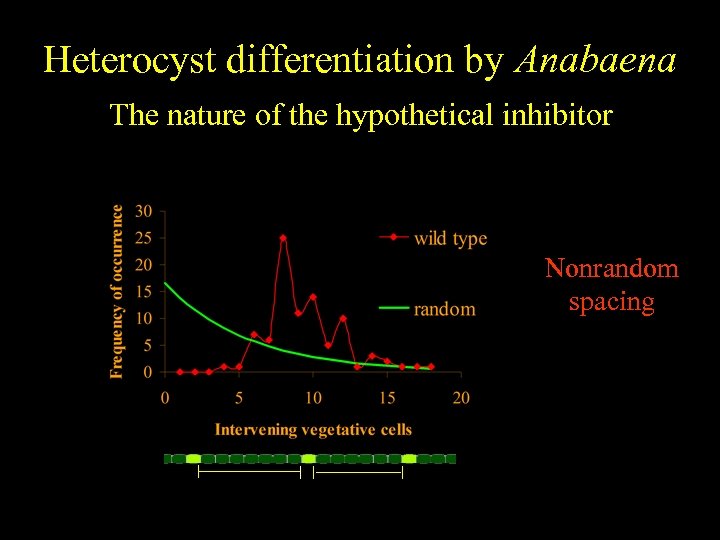 Heterocyst differentiation by Anabaena The nature of the hypothetical inhibitor Nonrandom spacing
Heterocyst differentiation by Anabaena The nature of the hypothetical inhibitor Nonrandom spacing
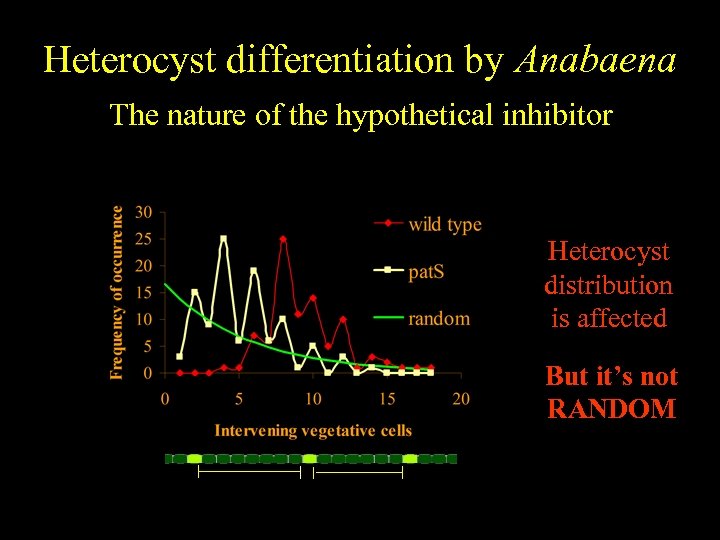 Heterocyst differentiation by Anabaena The nature of the hypothetical inhibitor Heterocyst distribution is affected But it’s not RANDOM
Heterocyst differentiation by Anabaena The nature of the hypothetical inhibitor Heterocyst distribution is affected But it’s not RANDOM
 Heterocyst differentiation by Anabaena A natural example of the Turing model? • Differentiation regulated by R-like protein, Het. R • Differentiation regulated by D-like protein, Pat. S • Pattern is not completely determined by Het. R and Pat. S
Heterocyst differentiation by Anabaena A natural example of the Turing model? • Differentiation regulated by R-like protein, Het. R • Differentiation regulated by D-like protein, Pat. S • Pattern is not completely determined by Het. R and Pat. S
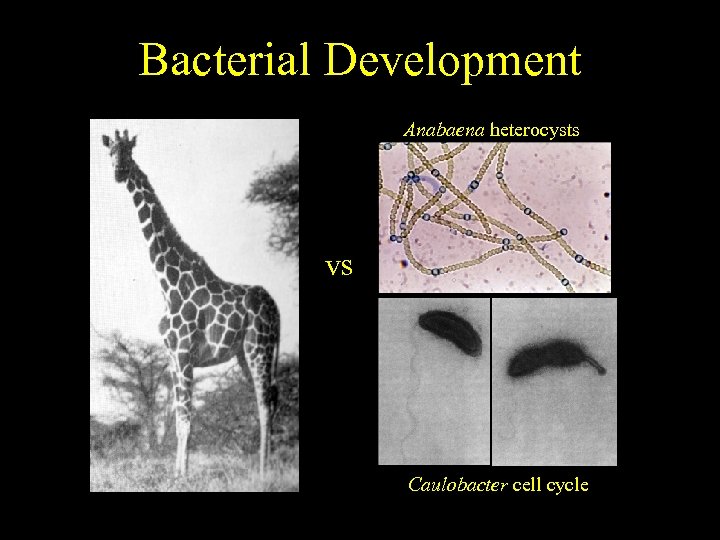 Bacterial Development End result. . . much simpler Bacillus sporulation Anabaena heterocysts vs Myxobacteria fruiting Caulobacter cell cycle
Bacterial Development End result. . . much simpler Bacillus sporulation Anabaena heterocysts vs Myxobacteria fruiting Caulobacter cell cycle
 How to understand complexity?
How to understand complexity?
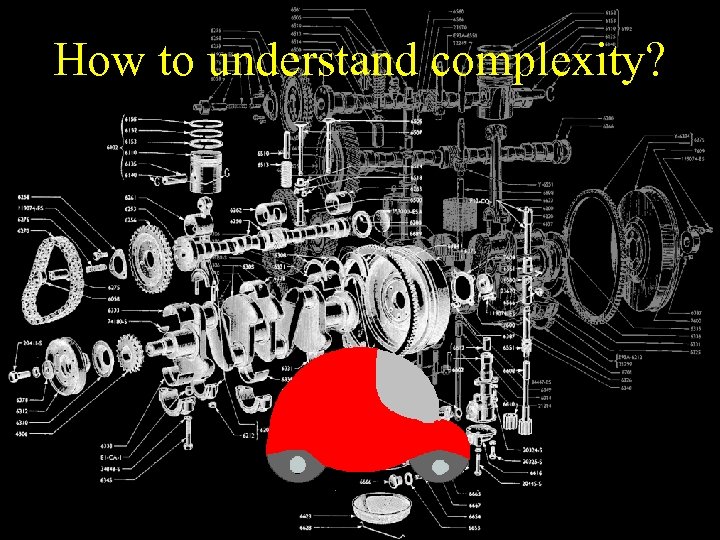 How to understand complexity?
How to understand complexity?



