77e1c3499d0b8f835bcbda223f869eaf.ppt
- Количество слайдов: 78
The CCPN Project Tim Stevens and Wayne Boucher October 2005

CCPN at Göteborg: Day 1 ■ Introduction to CCPN ■ The Ccp. Nmr applications ■ Analysis basics ■ Future developments ■ Analysis advanced
CCPN at Göteborg: Day 2 ■ An overview of the data model ■ API Tutorial ■ Analysis Macros ■ Widgets and Popups

CCPN Overview

The CCPN Project ■ Collaborative Computing Project for NMR ● Started in 1999 ● Collaborators in several countries ● Developers at University of Cambridge and EBI ■ Unifying platform for NMR software ● Similar to CCP 4 (X-ray) ■ Main goals: ● Data standards and data exchange ● Software development and distribution ● Meetings to determine and disseminate best practice ● Open source access

People ■ Cambridge ● Ernest Laue ● Rasmus Fogh ● Dan O’Donovan ■ EBI, Hinxton ● Kim Henrick ● John Ionides ● Wim Vranken ● Anne Pajon
History ■ Workshops: ● EBI (2000, 2001) ● Washington (2000) ■ Funding: ● BBSRC (2000 -2003, 2003 -2006) ● NMRQUAL (2001 -2004) ● TEMBLOR (2002 -2005) ● NMR-EXTEND (2005 -2008)
NMR Software ■ Problem - Heterogeneous development ● Lots of proprietary data formats ● Lots of stand-alone programs ● Data is ‘lost’ along the way ● Dedicated converters needed ● Not acceptable for structural genomics projects ■ Solution - Unity ● Data standards ■ Ease of transfer between programs ■ Completeness, integrity, deposition, data mining ● Libraries
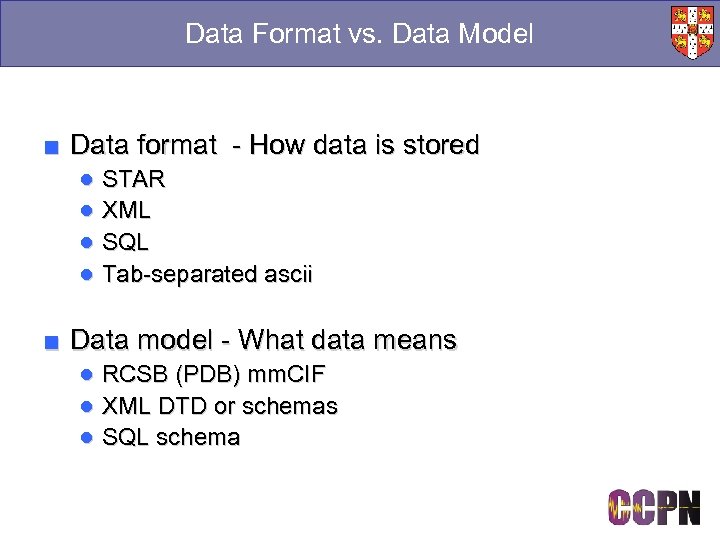
Data Format vs. Data Model ■ Data format - How data is stored ● STAR ● XML ● SQL ● Tab-separated ascii ■ Data model - What data means ● RCSB (PDB) mm. CIF ● XML DTD or schemas ● SQL schema
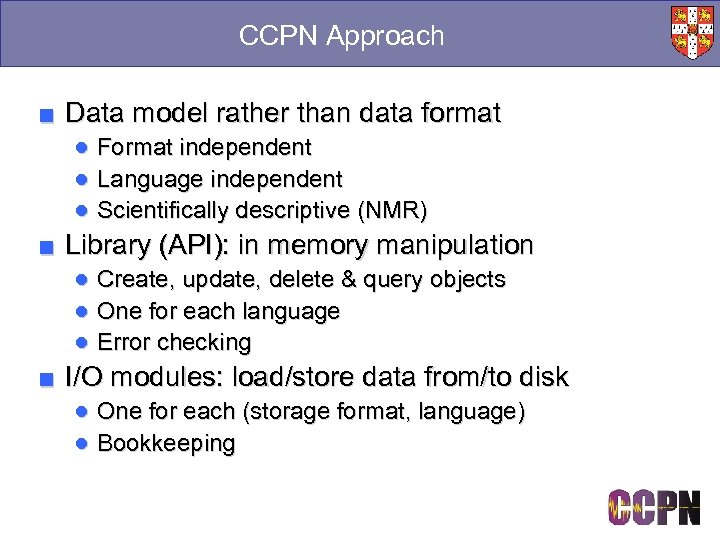
CCPN Approach ■ Data model rather than data format ● Format independent ● Language independent ● Scientifically descriptive (NMR) ■ Library (API): in memory manipulation ● Create, update, delete & query objects ● One for each language ● Error checking ■ I/O modules: load/store data from/to disk ● One for each (storage format, language) ● Bookkeeping
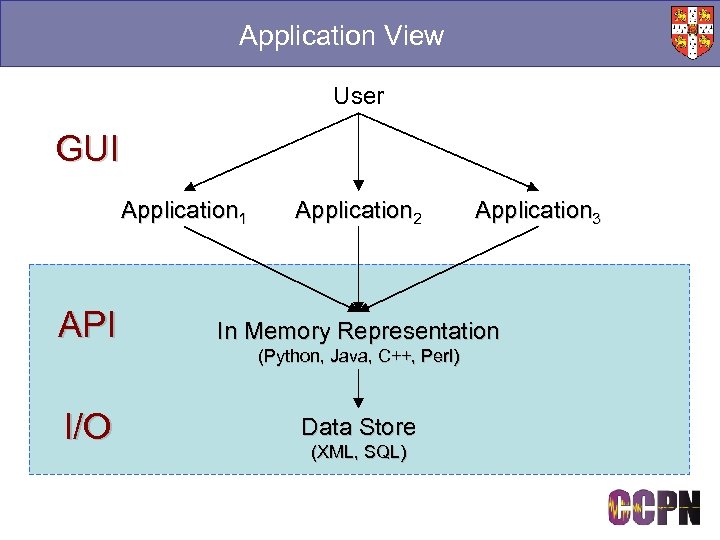
Application View User GUI Application 1 Application 2 Application 3 API In Memory Representation I/O Data Store (Python, Java, C++, Perl) (XML, SQL)
Model-Driven Architecture ■ UML: Unified Modelling Language ● Abstract representation of semantics ● Pictorial ■ Mapping from UML: to anything ● Multi-language ● Multi-format ● Architecture neutral (e. g. distributed or not) ■ Power: good and bad ■ CCPN uses Object Domain as its UML tool ● Python as scripting language
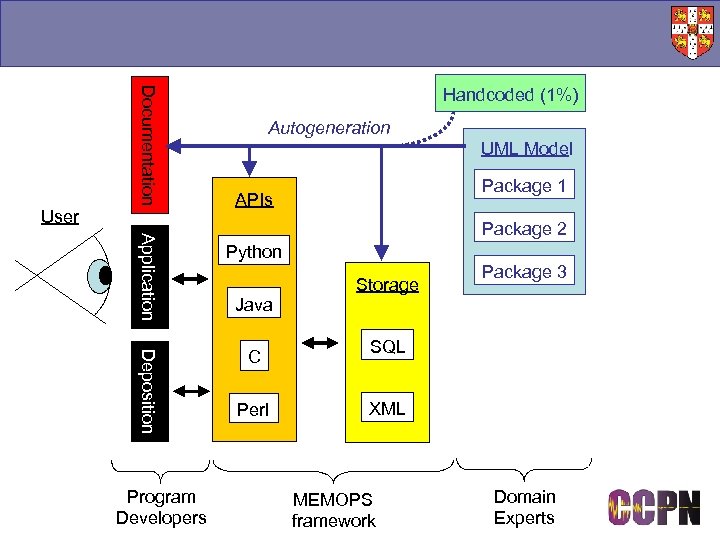
Documentation User Handcoded (1%) Autogeneration UML Model Package 1 APIs Application Python Deposition Package 2 C Program Developers Java Perl Storage Package 3 SQL XML MEMOPS framework Domain Experts
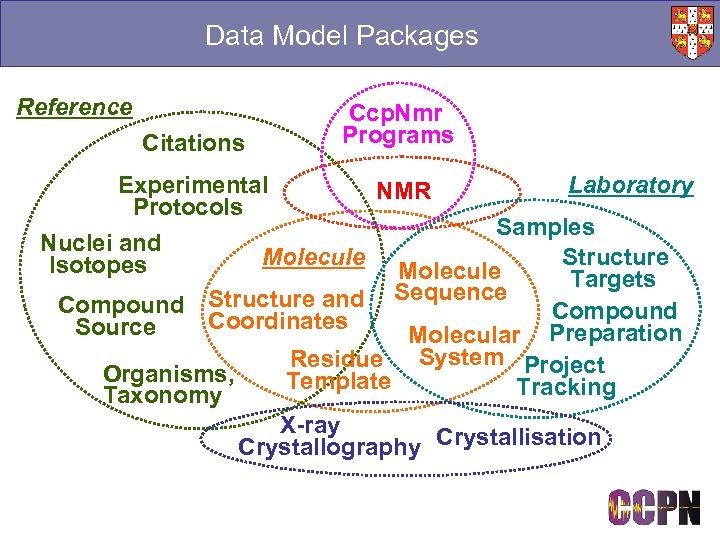
Data Model Packages Reference Citations Ccp. Nmr Programs Experimental Laboratory NMR Protocols Samples Nuclei and Molecule Structure Isotopes Molecule Targets Sequence Compound Structure and Compound Coordinates Source Molecular Preparation Residue System Project Organisms, Template Tracking Taxonomy X-ray Crystallography Crystallisation
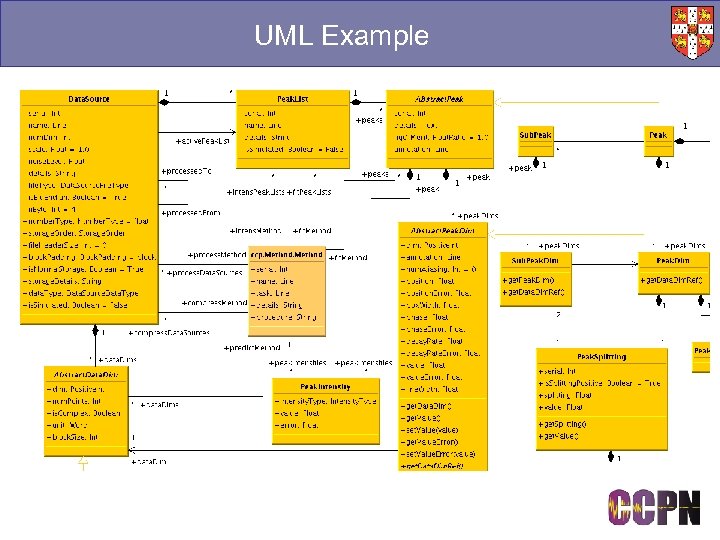
UML Example
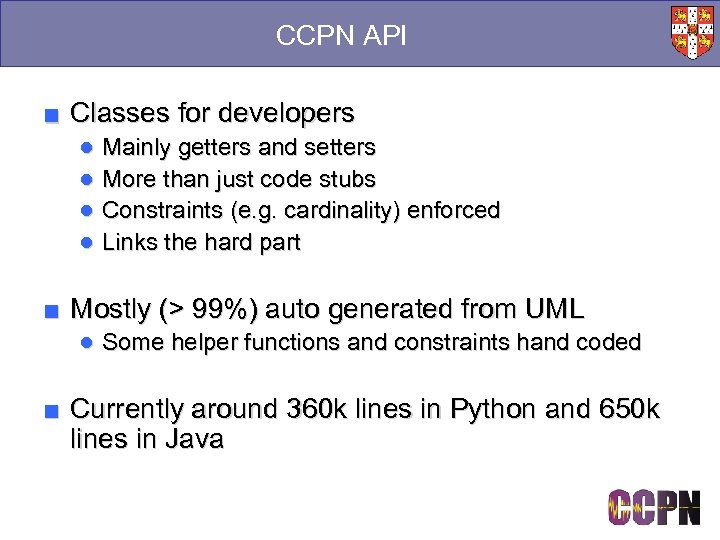
CCPN API ■ Classes for developers ● Mainly getters and setters ● More than just code stubs ● Constraints (e. g. cardinality) enforced ● Links the hard part ■ Mostly (> 99%) auto generated from UML ● Some helper functions and constraints hand coded ■ Currently around 360 k lines in Python and 650 k lines in Java
Developer Benefits ■ ■ Specified data model and API No I/O code Concentrate on science, not bookkeeping Extendible ● Application data can be assigned to any object ● UML model can be extended (packages) ■ Notification system ● Register interest when specified attribute changes (class, not object, level) ■ Undo/Redo (in future)
Current Status of API ■ Stable and released: ● Python and XML code generation ● NMR, molecule description and structure data model ■ In testing stages: ● Java and SQL database code generation ● Protein production data model ■ Preliminary: ● X-ray crystallography data model

Ccp. Nmr Applications
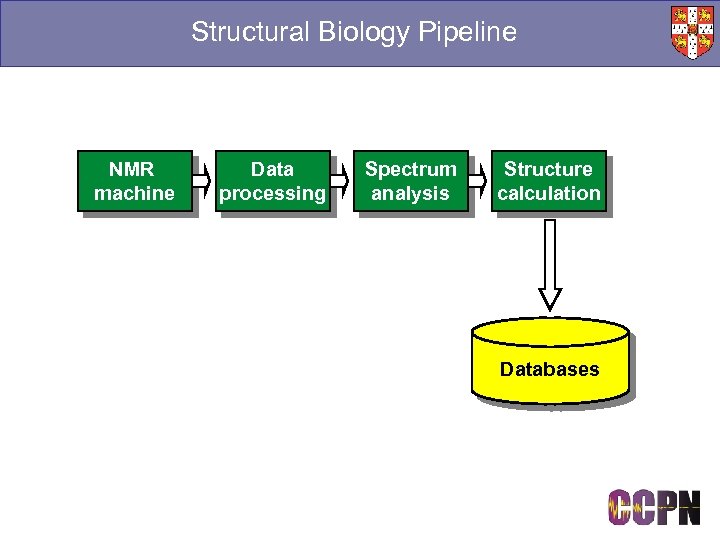
Structural Biology Pipeline NMR machine Data processing Spectrum analysis Structure calculation Databases
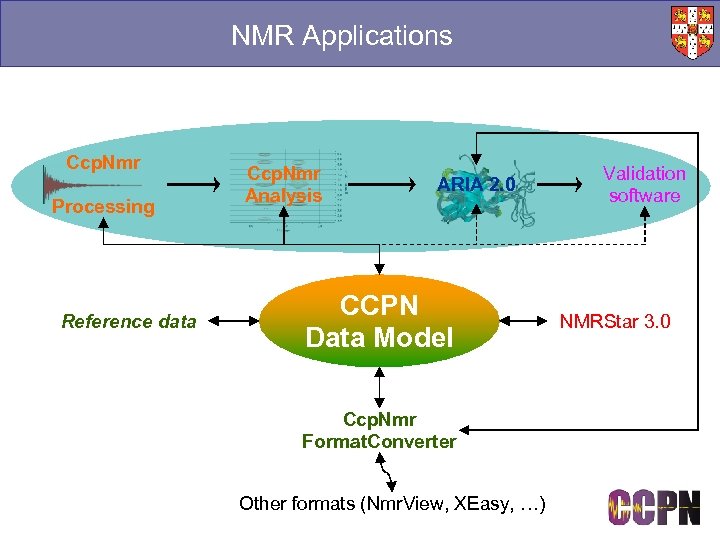
NMR Applications Ccp. Nmr Processing Reference data Ccp. Nmr Analysis ARIA 2. 0 CCPN Data Model Ccp. Nmr Format. Converter Other formats (Nmr. View, XEasy, …) Validation software NMRStar 3. 0
Main Ccp. Nmr Applications ■ Format Converter ● Conversion to and from legacy formats ■ Analysis ● Graphical analysis (e. g. assignment) program ■ Processing (coming soon) ● Azara “process” wrapped in data model
Ccp. Nmr Format Converter ■ Import/export of data formats to the Data Model ● For harvesting/deposition purposes ● Allow people to use or try out the data model ● Interaction with existing programs ■ Fully or partially handles: ● Ansig, Auremol, Autoassign, Azara, Bruker, Charmm, CNS/XPLOR/ARIA, Concoord, Diana/Dyana/Cyana, Discover, Fasta, Felix, Module, . mol, Molmol, Monte, Nmr. Draw, NMRPipe, NMR-STAR (v 2. 1. 1, v 3. 0), Nmr. View, Pdb, Pipp, Pistachio, Pronto, Sparky, Talos, Varian, XEasy ● Sequences, chemical compounds, coordinates, NMR measurements, constraints and peak lists, processing and acquisition parameters.
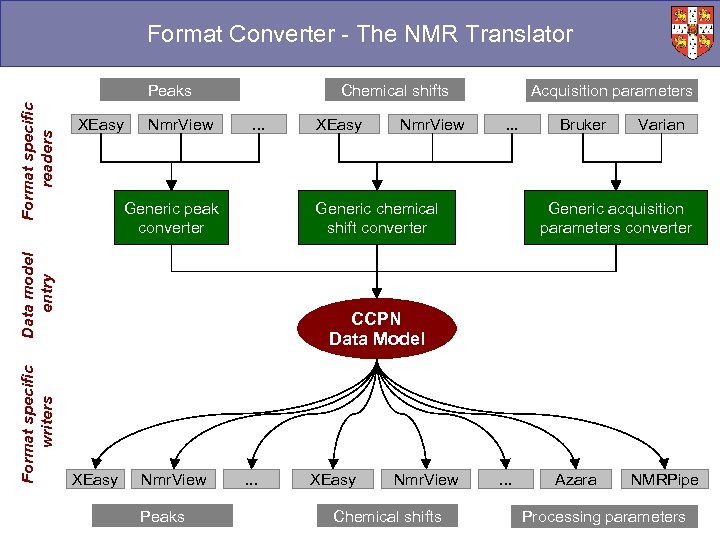
Format Converter - The NMR Translator XEasy Nmr. View Chemical shifts. . . Generic peak converter Format specific writers XEasy Nmr. View Acquisition parameters. . . Generic chemical shift converter Data model entry Format specific readers Peaks Bruker Varian Generic acquisition parameters converter CCPN Data Model XEasy Nmr. View Peaks . . . XEasy Nmr. View Chemical shifts . . . Azara NMRPipe Processing parameters
Format Converter Design ■ ■ ■ Wim Vranken (EBI) Set of Python scripts Accessed via: ● Tkinter (Tcl/Tk) ● custom Python scripts ■ http: //www. ebi. ac. uk/msdsrv/docs/NMRtoolkit/main. html
Ccp. Nmr Analysis ■ Requirements ● Cross platform ● Scalable ● Extensible ● Open and easy scripting language ● Modern graphical user interface ● Uses CCPN data model and API ■ Software ● Python, Tcl/Tk, C, Open. GL ● (Java, X, Motif) ■ OS ● Linux, Sun, SGI, OSX (Windows)
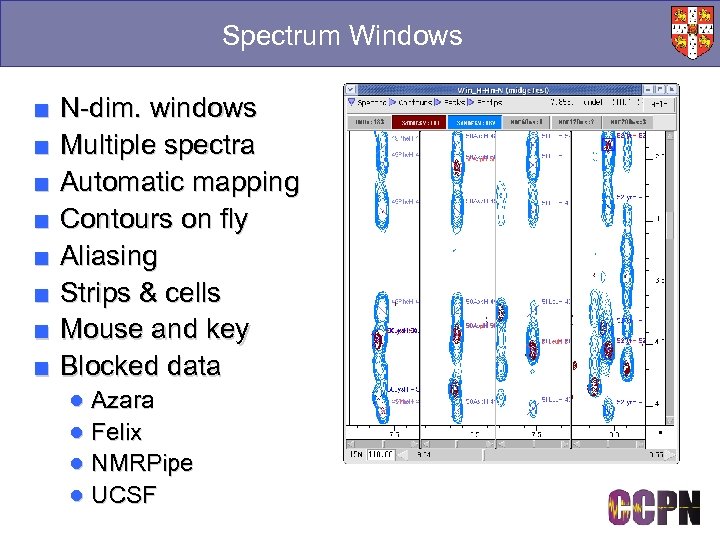
Spectrum Windows ■ ■ ■ ■ N-dim. windows Multiple spectra Automatic mapping Contours on fly Aliasing Strips & cells Mouse and key Blocked data ● Azara ● Felix ● NMRPipe ● UCSF
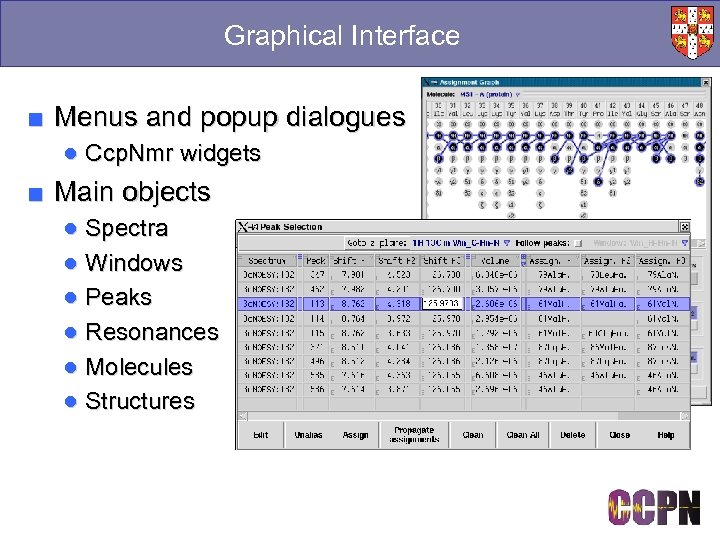
Graphical Interface ■ Menus and popup dialogues ● Ccp. Nmr widgets ■ Main objects ● Spectra ● Windows ● Peaks ● Resonances ● Molecules ● Structures
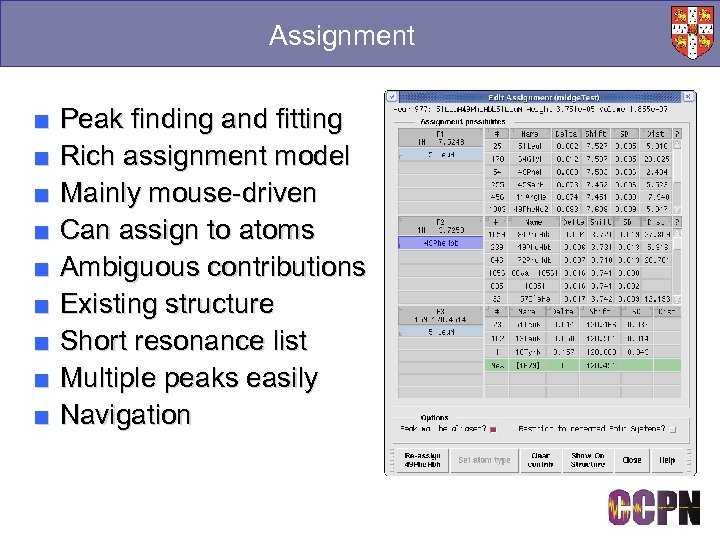
Assignment ■ ■ ■ ■ ■ Peak finding and fitting Rich assignment model Mainly mouse-driven Can assign to atoms Ambiguous contributions Existing structure Short resonance list Multiple peaks easily Navigation
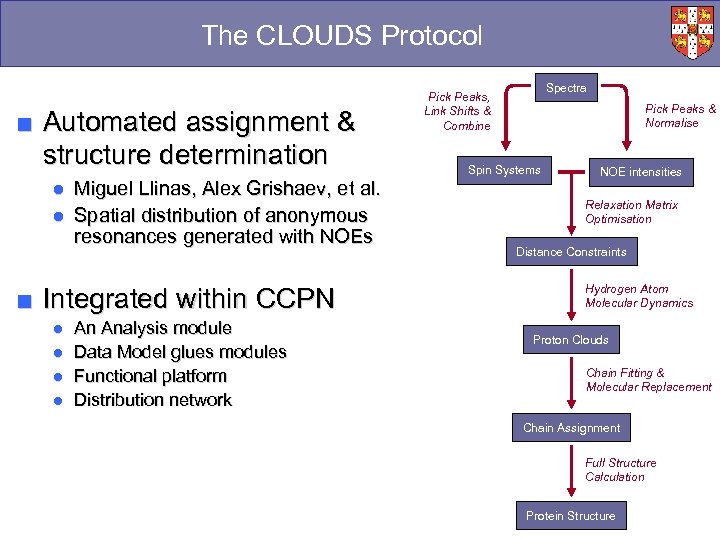
The CLOUDS Protocol ■ Automated assignment & structure determination ● Miguel Llinas, Alex Grishaev, et al. ● Spatial distribution of anonymous resonances generated with NOEs ■ Integrated within CCPN ● ● An Analysis module Data Model glues modules Functional platform Distribution network Spectra Pick Peaks, Link Shifts & Combine Pick Peaks & Normalise Spin Systems NOE intensities Relaxation Matrix Optimisation Distance Constraints Hydrogen Atom Molecular Dynamics Proton Clouds Chain Fitting & Molecular Replacement Chain Assignment Full Structure Calculation Protein Structure
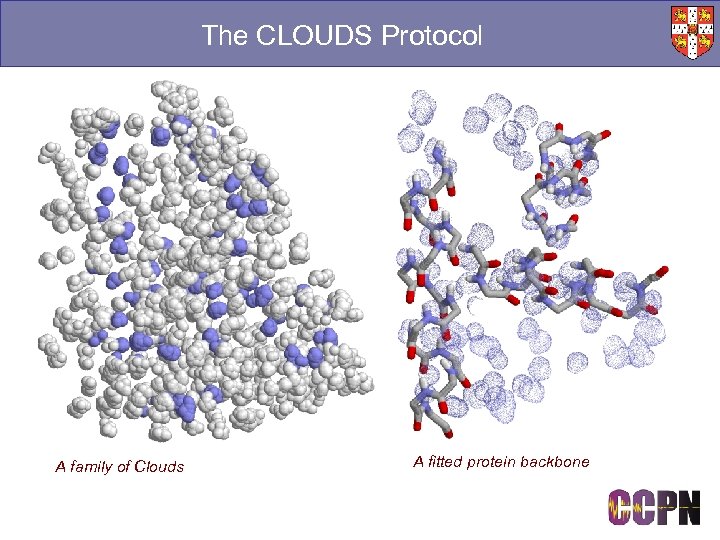
The CLOUDS Protocol A family of Clouds A fitted protein backbone
Other Features ■ ■ Works with Format. Converter Chemical compounds database NMR reference information Hard copy ● Post. Script ● PDF ■ ■ Table export Rate analysis Macros Structures
Ccp. Nmr Analysis Tutorial Part I

CCPN Future
Extend-NMR ■ EU STREP application funded to fully integrate software from: ● Bruker (TOPSPIN, acquisition) ● Billeter, Orekhov (Garant, Munin, MDD) ● Kalbitzer (Auremol) ● Llinas (CLOUDS) ● Nilges (Inferential Structure Determination) ● Bonvin (Haddock, RECOORD) ● Vriend, Vuister (Queen, What-Check) ● Henrick, Vranken (NMR database) ■ Focus on complexes and development of better software methodology
LIMS Collaborations ■ PIMS project collaboration ● Protein production LIMS (with EBI, Sport Consortia, OPPF and Poupon) ■ EU STREP application (SFGLIMS) to work with : ● Poupon (Protein Production) ● Perrakis (Biophysical methods, crystallisation) ● Bricogne (X-ray data collection and structure generation) ● Prilusky, Sussman (Bioinformatics, data mining)
Data Model Extensions ■ EXTEND-NMR ● New NMR applications ■ ■ Solid state NMR PIMS ● LIMS for protein production ■ SFGLIMS ● LIMS for NMR and X-ray structure determination ■ ■ ■ X-ray Chemoinformatics (Metabolomics? )
Code Generation Plans ■ C++/C/FORTRAN code ● Needed for Extend-NMR and for Ccp. Nmr Processing ● Needed for interface to CYANA, NMRPIPE, AUTOPSY, etc. ■ Java/Database code ● Extend for LIMS, high-throughput projects, NMRVIEW ■ Basic Machinery ● Upgrades for long term extensibility/maintainability and performance
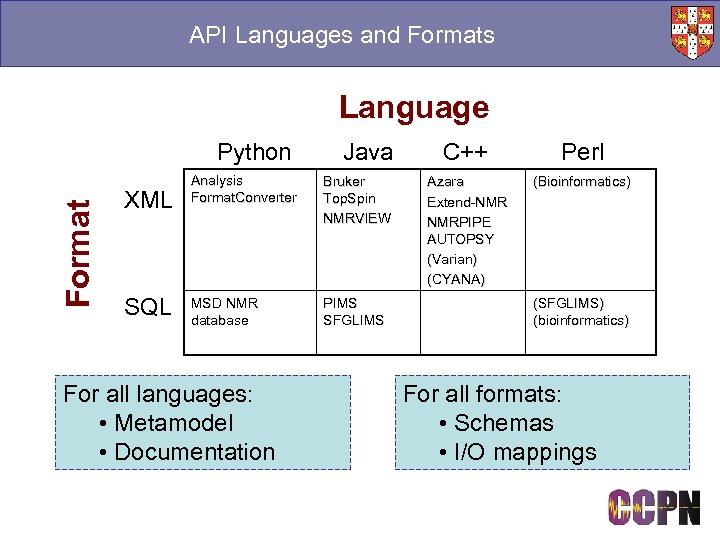
API Languages and Formats Language Format Python XML SQL Java Analysis Format. Converter Bruker Top. Spin NMRVIEW MSD NMR database PIMS SFGLIMS For all languages: • Metamodel • Documentation C++ Perl Azara Extend-NMR NMRPIPE AUTOPSY (Varian) (CYANA) (Bioinformatics) (SFGLIMS) (bioinformatics) For all formats: • Schemas • I/O mappings
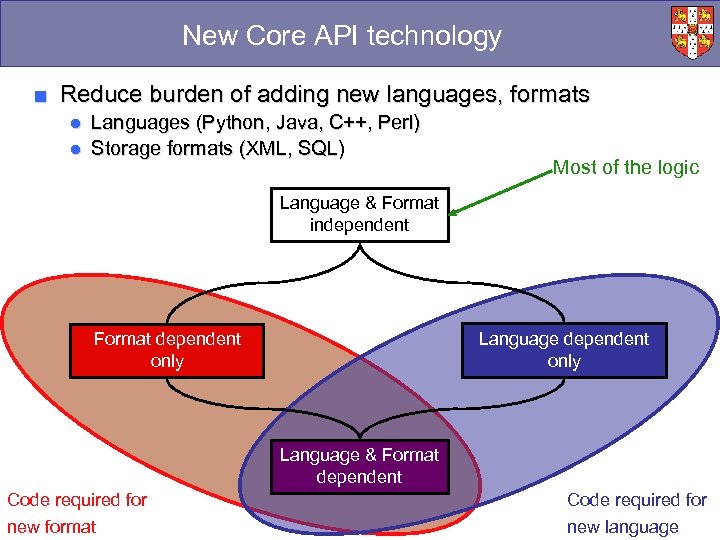
New Core API technology ■ Reduce burden of adding new languages, formats ● Languages (Python, Java, C++, Perl) ● Storage formats (XML, SQL) Most of the logic Language & Format independent Language dependent only Format dependent only Language & Format dependent Code required for new format new language
Core API technology, cont. ■ Remodelling of implementation details ● Storages, collection types, root objects, etc. ■ Complex data types ● e. g. rotation matrix ■ Client/Server architecture ● For PIMS and SFGLIMS
Analysis Development ■ Beyond CLOUDS ● Large proteins, homologues ■ Processing linked in ■ Couplings (RDCs, TROSY), dihedral constraints ■ Titrations (Ka, Kd) ■ Chain states (alternate conformations) ■ Solid State NMR ■ Organic chemistry NMR (1 D) ■ Publication-ready diagrams and tables ■ Windows version
Developments in Extend-NMR ■ Integrated Bayesian, maximum entropy, … methods for data-processing, analysis and structure calculation ■ ‘Molecular replacement’ for NMR ■ Further RECOORD development ■ Databank for Experimental NMR spectra (DEN) ■ MSD database analysis
Licenses ■ GPL ● Data model ● Scripts which produce APIs ■ LGPL ● Generic libraries ● Widget libraries ● Format Converter ■ CCPN ● Analysis

Resources, 1 ■ Source. Forge: ● CVS repository for code ● API and Format. Converter releases ● http: //sourceforge. net/projects/ccpn ■ CCPN: ● Meetings, workshops ● API, Format. Converter and Analysis releases ● http: //www. ccpn. ac. uk

Resources, 2 ■ EBI: ● Format Converter ● Databases (MSD group) ● http: //www. ebi. ac. uk/msdsrv/docs/NMRtoolkit/main. html ■ JISCMAIL: ● Email list ● http: //www. jiscmail. ac. uk/lists/ccpnmr. html ● (http: //www. jiscmail. ac. uk/lists/nmrgen. html)
Ccp. Nmr Analysis Tutorial Part II
CCPN at Göteborg: Day 2 ■ An overview of the data model ■ API Tutorial ■ Analysis Macros ■ Widgets and Popups
Major Data Model Packages
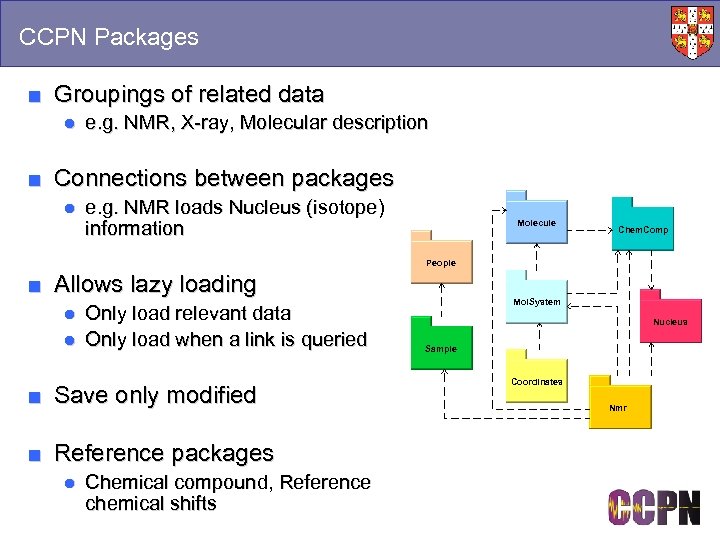
CCPN Packages ■ Groupings of related data ● e. g. NMR, X-ray, Molecular description ■ Connections between packages ● e. g. NMR loads Nucleus (isotope) information Molecule Chem. Comp People ■ Allows lazy loading ● Only load relevant data ● Only load when a link is queried ■ Save only modified ■ Reference packages ● Chemical compound, Reference chemical shifts Mol. System Nucleus Sample Coordinates Nmr
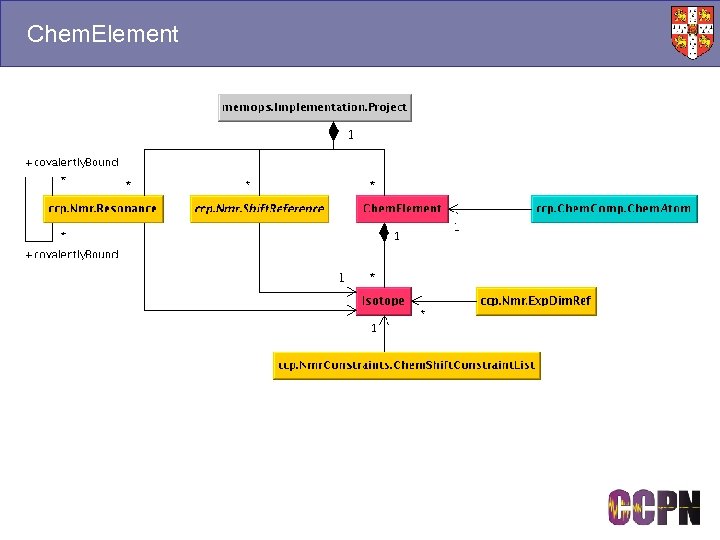
Chem. Element
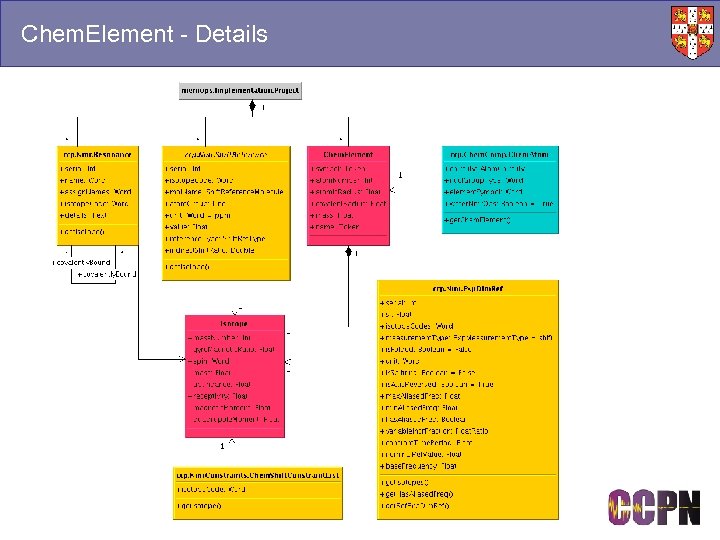
Chem. Element - Details
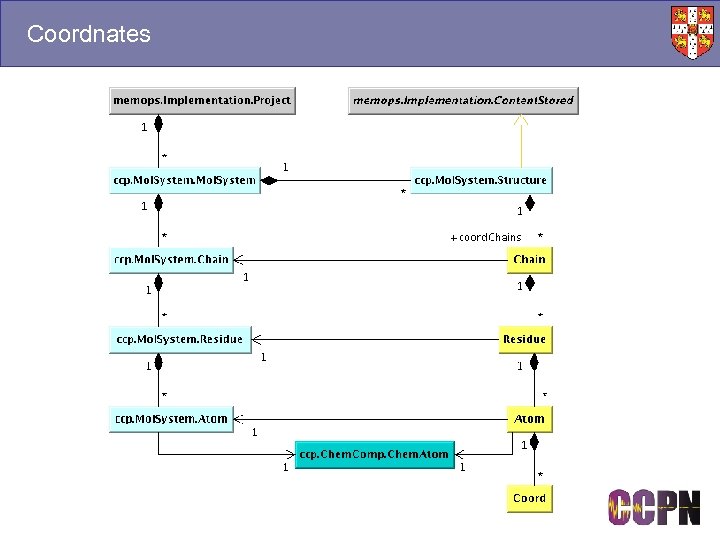
Coordnates
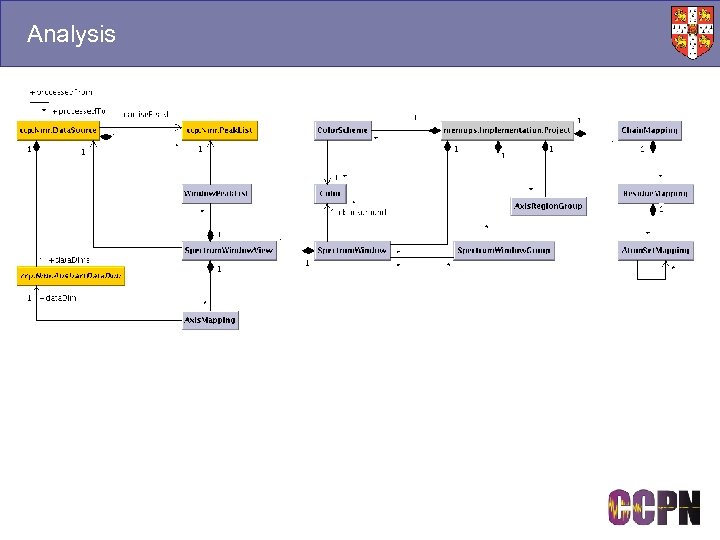
Analysis
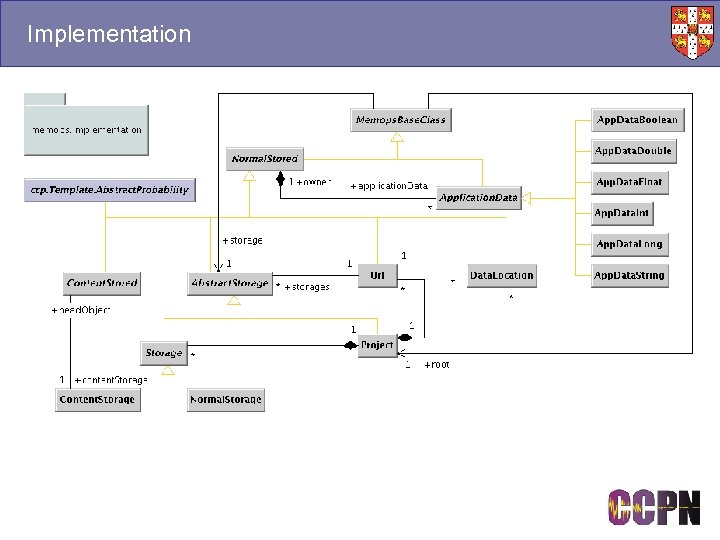
Implementation
Molecules and Mol. Systems ■ Molecules ● Templates for specifying molecular connectivity. ● Sequences, chemical components, protonation state etc. ● A kind of reference, e. g. “Lysozyme” ■ Mol. Systems ● Contain chains, which contain residues, which contain atoms. ● The objects you assign to. ● Built using molecule templates, e. g. a homo-oligomer is built using the same template to make different chains. ■ Stored in different packages ● Molecule. xml, Mol. System. xml
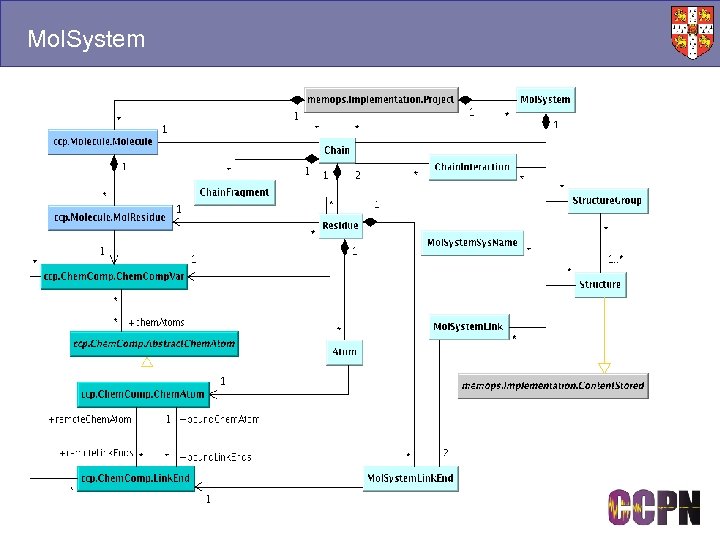
Mol. System
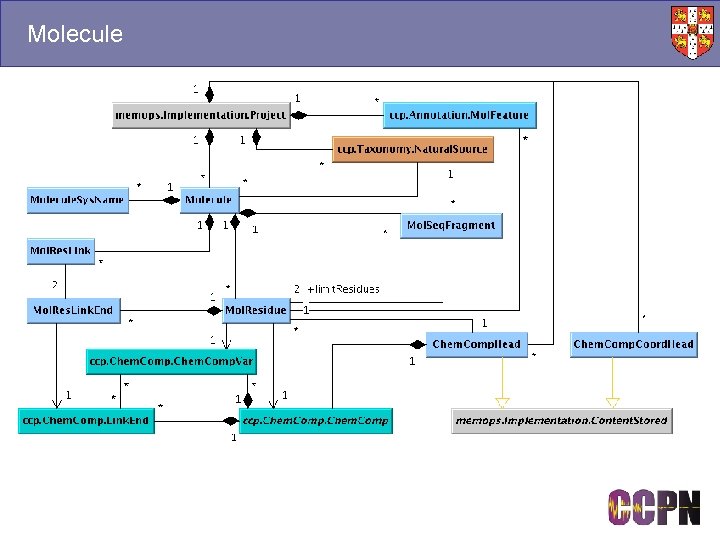
Molecule
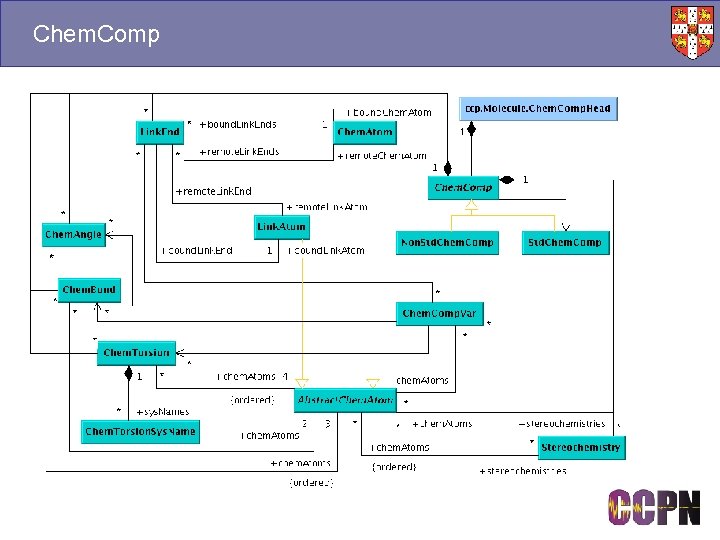
Chem. Comp
Experiment, Spectrum & Shift List Objects ■ Experiment ● The set-up under particular conditions at a particular time, not a class of experiment. ■ Spectrum ● Known as Data Source in the data model. A pointer to a chunk of data that results from an experiment. Several spectra may result from the same experiment if they are processed differently. ■ Peak List ● A set of crosspeaks that have been picked for a spectrum. A spectrum can have several peak lists. The user can separate peaks into classes, e. g. picked in different ways. ■ Shift List ● A set of chemical shifts, which are derived from peaks and may be linked to atoms. Valid for a set of experiments with similar conditions that give similar chemical shifts. Using different shift lists doesn’t change assignments, but it does change which peaks are used in the calculation of a shift value.
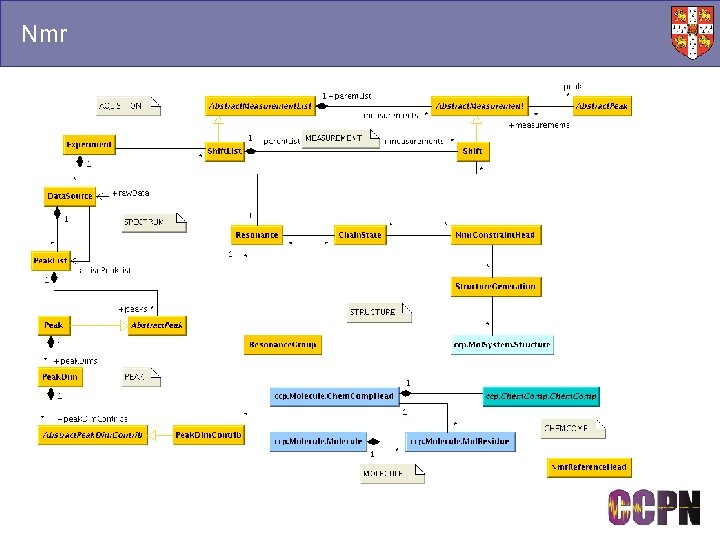
Nmr
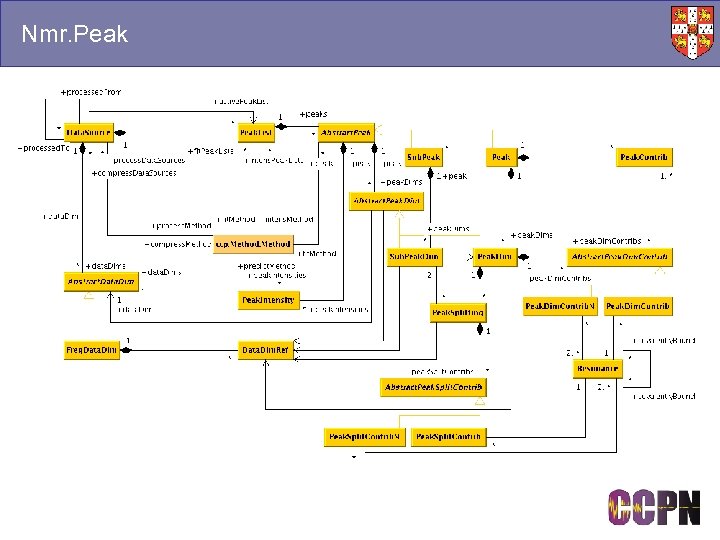
Nmr. Peak
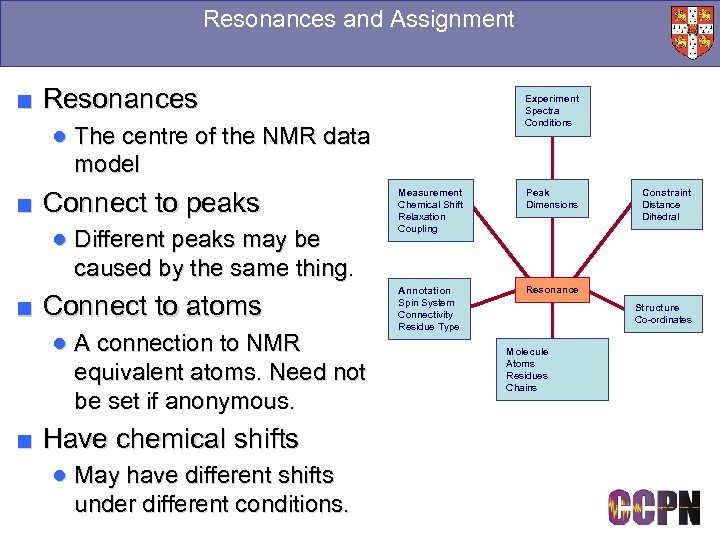
Resonances and Assignment ■ Resonances Experiment Spectra Conditions ● The centre of the NMR data model ■ Connect to peaks ● Different peaks may be caused by the same thing. ■ Connect to atoms ● A connection to NMR equivalent atoms. Need not be set if anonymous. ■ Have chemical shifts ● May have different shifts under different conditions. Measurement Chemical Shift Relaxation Coupling Peak Dimensions Annotation Spin System Connectivity Residue Type Constraint Distance Dihedral Resonance Structure Co-ordinates Molecule Atoms Residues Chains
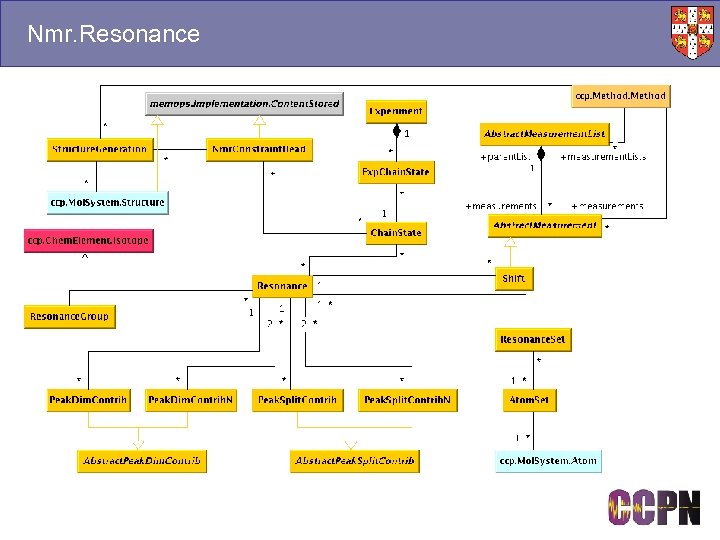
Nmr. Resonance
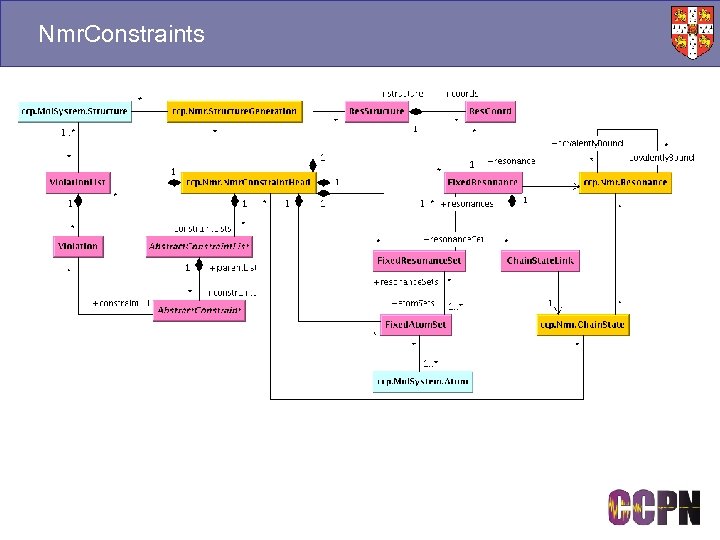
Nmr. Constraints
Python API coding tutorial
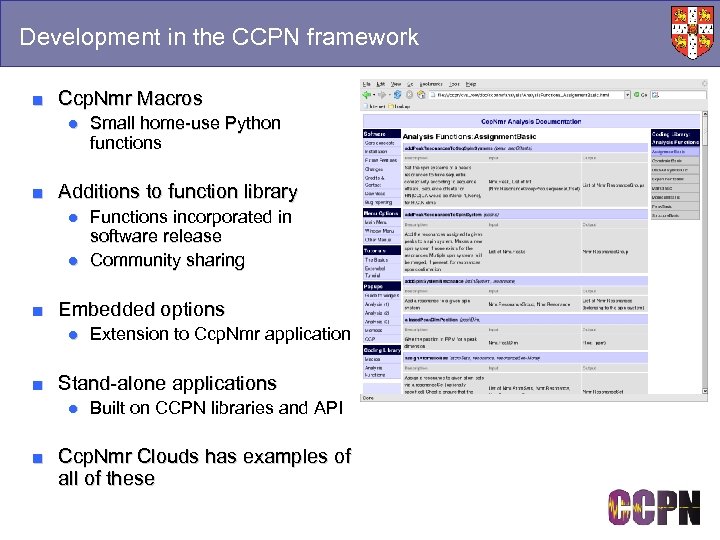
Development in the CCPN framework ■ Ccp. Nmr Macros ● Small home-use Python functions ■ Additions to function library ● Functions incorporated in software release ● Community sharing ■ Embedded options ● Extension to Ccp. Nmr application ■ Stand-alone applications ● Built on CCPN libraries and API ■ Ccp. Nmr Clouds has examples of all of these
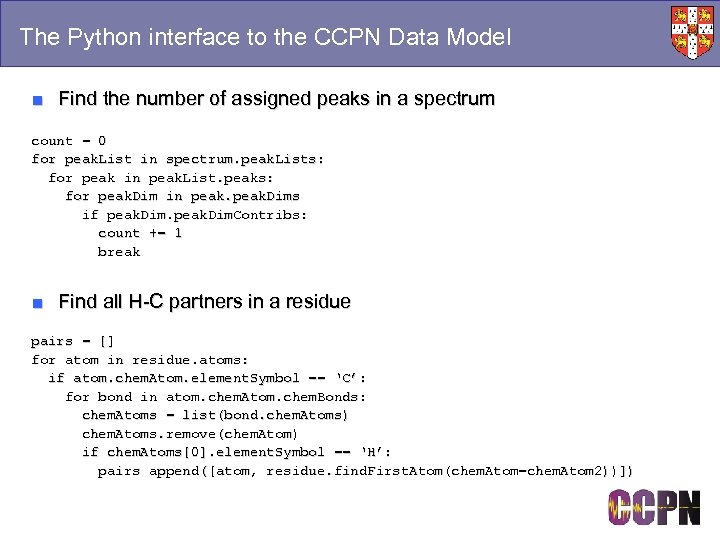
The Python interface to the CCPN Data Model ■ Find the number of assigned peaks in a spectrum count = 0 for peak. List in spectrum. peak. Lists: for peak in peak. List. peaks: for peak. Dim in peak. Dims if peak. Dim. Contribs: count += 1 break ■ Find all H-C partners in a residue pairs = [] for atom in residue. atoms: if atom. chem. Atom. element. Symbol == ‘C’: for bond in atom. chem. Atom. chem. Bonds: chem. Atoms = list(bond. chem. Atoms) chem. Atoms. remove(chem. Atom) if chem. Atoms[0]. element. Symbol == ‘H’: pairs append([atom, residue. find. First. Atom(chem. Atom=chem. Atom 2))])
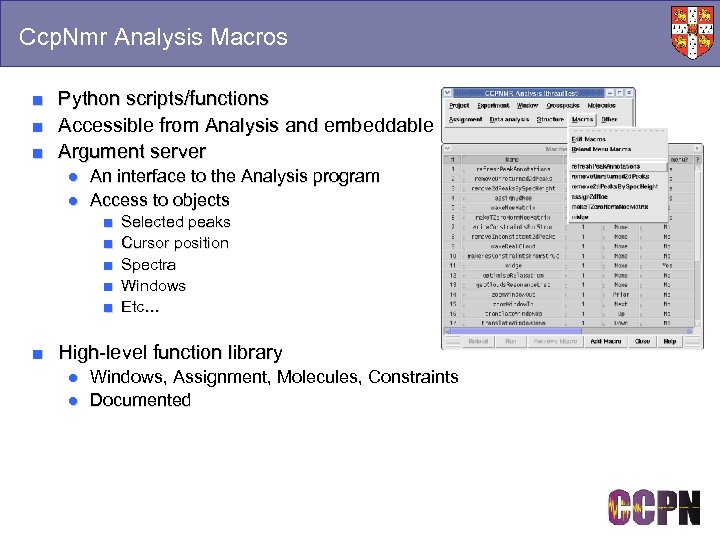
Ccp. Nmr Analysis Macros ■ ■ ■ Python scripts/functions Accessible from Analysis and embeddable Argument server ● An interface to the Analysis program ● Access to objects ■ ■ ■ Selected peaks Cursor position Spectra Windows Etc… ■ High-level function library ● Windows, Assignment, Molecules, Constraints ● Documented

Macro 1 - Simple stuff • Python language • Function anatomy • Import library functions • Argument. Server • Simple program def add. Marks. To. Peaks(arg. Server, peaks=None): """Descrn: Adds position line markers to the selected peaks. Inputs: Argument. Server, List of Nmr. Peaks Output: None """ from ccpnmr. analysis. Mark. Basic import create. Peak. Mark if not peaks: peaks = arg. Server. get. Current. Peaks() # no peaks - nothing happens for peak in peaks: create. Peak. Mark(peak, remove=0)

Macro 2 - Ask the user def calc. Average. Peak. List. Intensity(arg. Server, peak. List=None, intensity. Type='height'): """Descrn: Find the average height of peaks in a peak list. Inputs: Argument. Server, Nmr. Peak. List Output: Float """ from ccpnmr. analysis. Constraint. Basic import get. Mean. Peak. Intensity if not peak. List: peak. List = arg. Server. get. Peak. List() if not peak. List: arg. Server. show. Warning('No peak list selected') return answer = arg. Server. ask. Yes. No('Use peak volumes? Height will be used otherwise. ') if answer: # is true intensity. Type = 'volume' spec expt intensity data = = peak. List. data. Source spec. experiment get. Mean. Peak. Intensity(peak. List. peaks, intensity. Type=intensity. Type) (intensity. Type, expt. name, spec. name, peak. List. serial, intensity )) arg. Server. show. Info('Mean peak %s for %s %s peak list %d is %e' % data return intensity
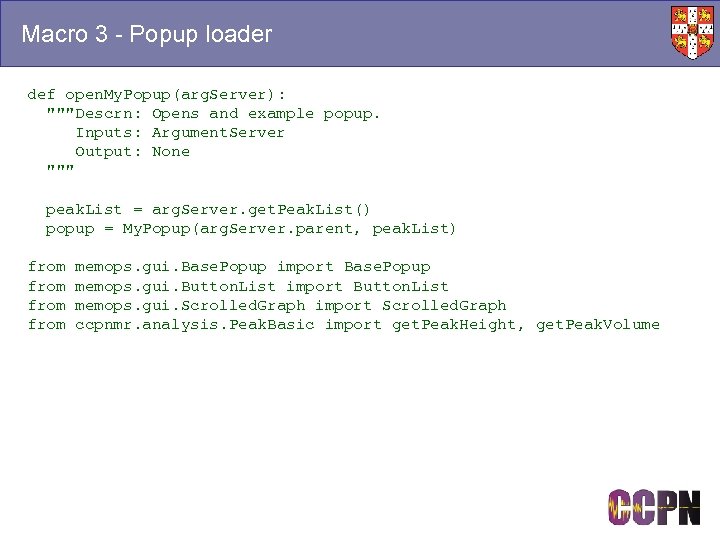
Macro 3 - Popup loader def open. My. Popup(arg. Server): """Descrn: Opens and example popup. Inputs: Argument. Server Output: None """ peak. List = arg. Server. get. Peak. List() popup = My. Popup(arg. Server. parent, peak. List) from memops. gui. Base. Popup import Base. Popup memops. gui. Button. List import Button. List memops. gui. Scrolled. Graph import Scrolled. Graph ccpnmr. analysis. Peak. Basic import get. Peak. Height, get. Peak. Volume

Macro 3 - The popup class My. Popup(Base. Popup): def __init__(self, parent, peak. List, *args, **kw): self. peak. List = peak. List self. colours = ['red', 'green'] self. data. Sets = [] Base. Popup. __init__(self, parent=parent, title='Test Popup', **kw) def body(self, gui. Parent): row = 0 self. graph = Scrolled. Graph(gui. Parent) self. graph. grid(row=row, column=0, sticky='NSEW') row += 1 texts = ['Draw graph', 'Goodbye'] commands = [self. draw, self. destroy] buttons = Button. List(gui. Parent, texts=texts, commands = commands) buttons. grid(row=row, column=0, sticky='NSEW') def draw(self): self. data. Sets = self. get. Data() self. graph. update(self. data. Sets, self. colours) def get. Data(self): peak. Data = [( get. Peak. Volume(peak) or 0. 0, peak) for peak in self. peak. List. peaks] peak. Data. sort() heights = [] volumes = [] i = 0 for volume, peak in peak. Data: heights. append([i, get. Peak. Height(peak) or 0. 0]) volumes. append([i, volume]) i += 1
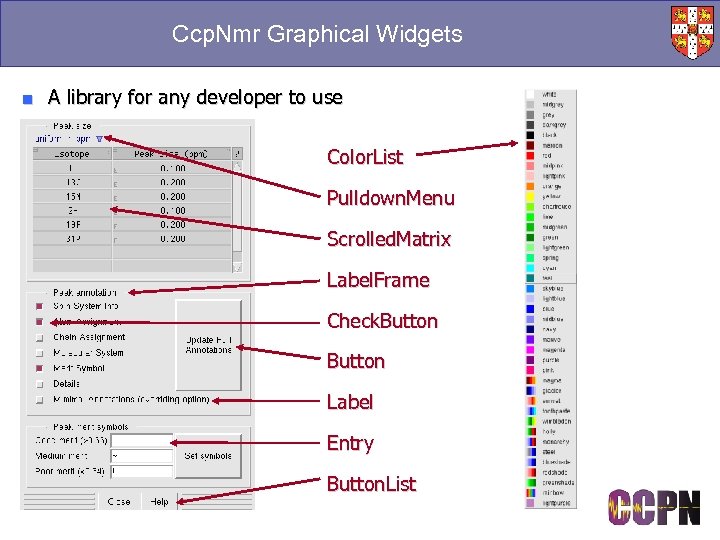
Ccp. Nmr Graphical Widgets ■ A library for any developer to use Color. List Pulldown. Menu Scrolled. Matrix Label. Frame Check. Button Label Entry Button. List
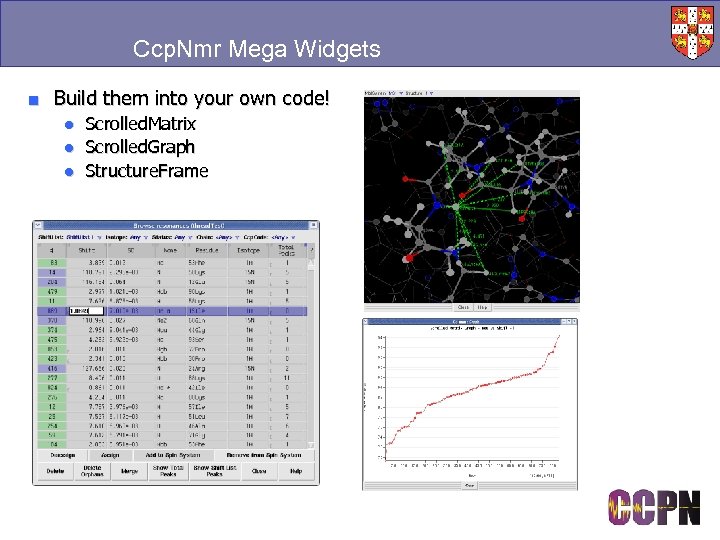
Ccp. Nmr Mega Widgets ■ Build them into your own code! ● Scrolled. Matrix ● Scrolled. Graph ● Structure. Frame
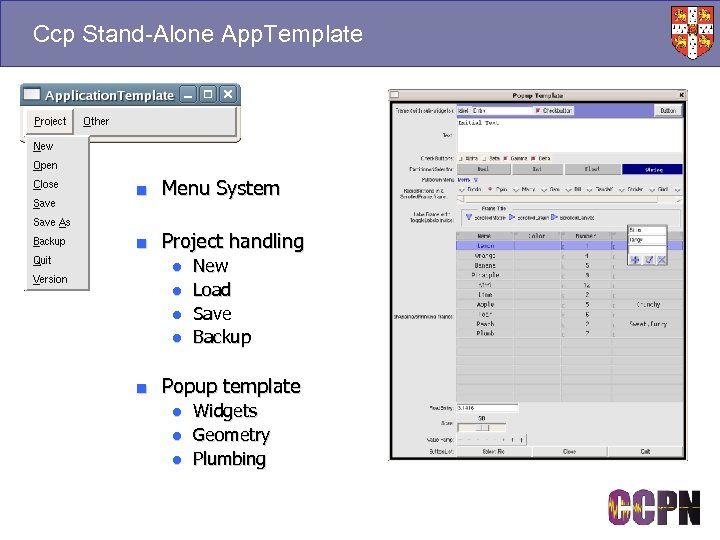
Ccp Stand-Alone App. Template ■ Menu System ■ Project handling ● ● New Load Save Backup ■ Popup template ● Widgets ● Geometry ● Plumbing
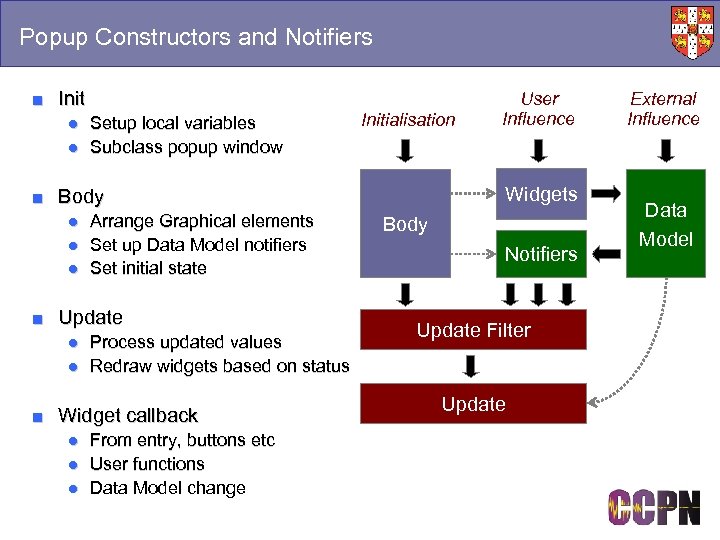
Popup Constructors and Notifiers ■ Init ● Setup local variables ● Subclass popup window Initialisation Widgets ■ Body ● Arrange Graphical elements ● Set up Data Model notifiers ● Set initial state ■ Update ● Process updated values ● Redraw widgets based on status ■ Widget callback ● From entry, buttons etc ● User functions ● Data Model change User Influence Body Notifiers Update Filter Update External Influence Data Model

Aftercare ■ www. ccpn. ac. uk ● Downloads ● Data Model documentation ● Analysis documentation ● Tutorials ■ Mailing List ● http: //www. jiscmail. ac. uk/lists/CCPNMR. html ● Quick response ● Bugs ● Requests
77e1c3499d0b8f835bcbda223f869eaf.ppt