ff80addbf5f1d84a08f26ccdb3a15abf.ppt
- Количество слайдов: 32
 The application of real-time PCR techniques in the diagnosis of respiratory infections. Dr. Dirk Vollenbroich Minerva Biolabs Gmb. H
The application of real-time PCR techniques in the diagnosis of respiratory infections. Dr. Dirk Vollenbroich Minerva Biolabs Gmb. H
 Mycoplasma pneumoniae • prevalence of 1 to 30 %, only Streptococcus pneumoniae is more frequent • frequent cause of community-acquired respiratory infection in children and adults • atypical Community Acquired Pneumonia • mostly outbreaks in institutional settings, extensive epidemics in Europe every 4 to 5 years • 2 subtypes and 2 variants are known
Mycoplasma pneumoniae • prevalence of 1 to 30 %, only Streptococcus pneumoniae is more frequent • frequent cause of community-acquired respiratory infection in children and adults • atypical Community Acquired Pneumonia • mostly outbreaks in institutional settings, extensive epidemics in Europe every 4 to 5 years • 2 subtypes and 2 variants are known
 Legionella • rare in Northern Europe, mediteranian countries 8 - 22 % • direct infection through aerosoles • clinic: Pontiac fever, Legionairs´ disease • more than 62 species, 73 serogroups • at least 18 of these described as pathogenic • current distribution: 50 -70 % Legionella pneumophila SG 1 20 -30 % Legionella pneumophila SG 2 - 14 5 -10 % Legionella species
Legionella • rare in Northern Europe, mediteranian countries 8 - 22 % • direct infection through aerosoles • clinic: Pontiac fever, Legionairs´ disease • more than 62 species, 73 serogroups • at least 18 of these described as pathogenic • current distribution: 50 -70 % Legionella pneumophila SG 1 20 -30 % Legionella pneumophila SG 2 - 14 5 -10 % Legionella species
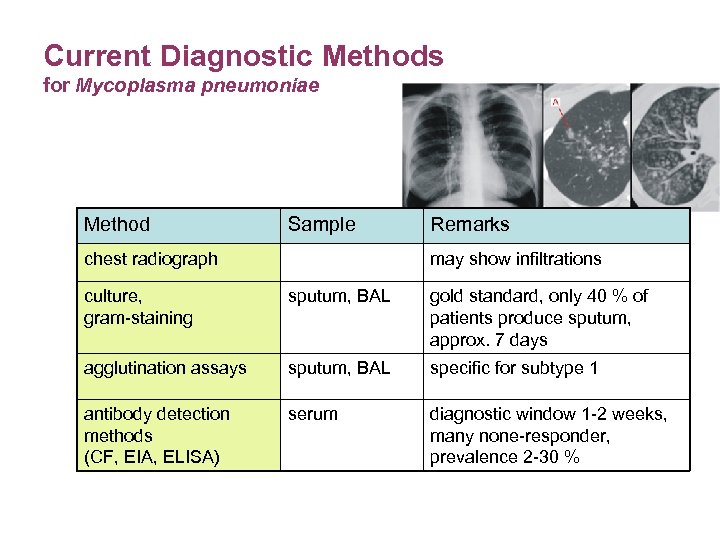 Current Diagnostic Methods for Mycoplasma pneumoniae Method Sample chest radiograph Remarks may show infiltrations culture, gram-staining sputum, BAL gold standard, only 40 % of patients produce sputum, approx. 7 days agglutination assays sputum, BAL specific for subtype 1 antibody detection methods (CF, EIA, ELISA) serum diagnostic window 1 -2 weeks, many none-responder, prevalence 2 -30 %
Current Diagnostic Methods for Mycoplasma pneumoniae Method Sample chest radiograph Remarks may show infiltrations culture, gram-staining sputum, BAL gold standard, only 40 % of patients produce sputum, approx. 7 days agglutination assays sputum, BAL specific for subtype 1 antibody detection methods (CF, EIA, ELISA) serum diagnostic window 1 -2 weeks, many none-responder, prevalence 2 -30 %
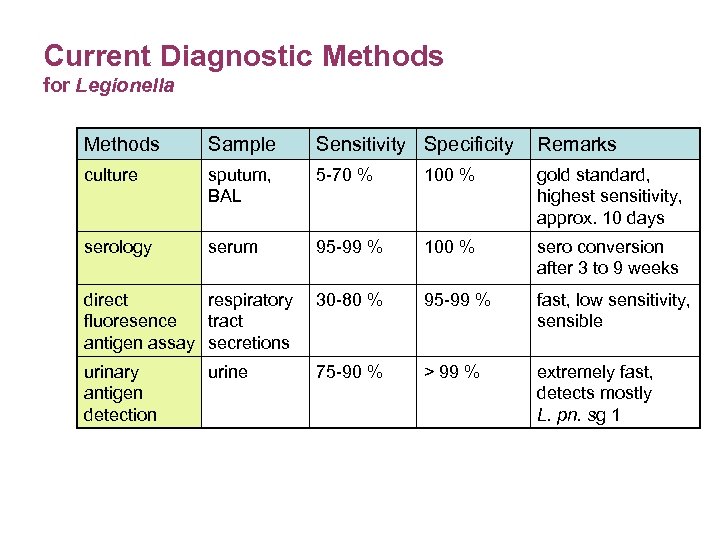 Current Diagnostic Methods for Legionella Methods Sample Sensitivity Specificity Remarks culture sputum, BAL 5 -70 % 100 % gold standard, highest sensitivity, approx. 10 days serology serum 95 -99 % 100 % sero conversion after 3 to 9 weeks direct respiratory fluoresence tract antigen assay secretions 30 -80 % 95 -99 % fast, low sensitivity, sensible urinary antigen detection 75 -90 % > 99 % extremely fast, detects mostly L. pn. sg 1 urine
Current Diagnostic Methods for Legionella Methods Sample Sensitivity Specificity Remarks culture sputum, BAL 5 -70 % 100 % gold standard, highest sensitivity, approx. 10 days serology serum 95 -99 % 100 % sero conversion after 3 to 9 weeks direct respiratory fluoresence tract antigen assay secretions 30 -80 % 95 -99 % fast, low sensitivity, sensible urinary antigen detection 75 -90 % > 99 % extremely fast, detects mostly L. pn. sg 1 urine
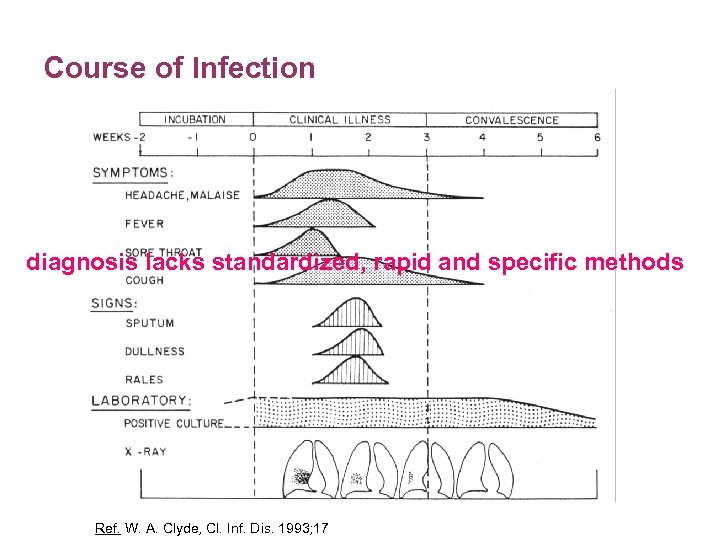 Course of Infection diagnosis lacks standardized, rapid and specific methods Ref. W. A. Clyde, Cl. Inf. Dis. 1993; 17
Course of Infection diagnosis lacks standardized, rapid and specific methods Ref. W. A. Clyde, Cl. Inf. Dis. 1993; 17
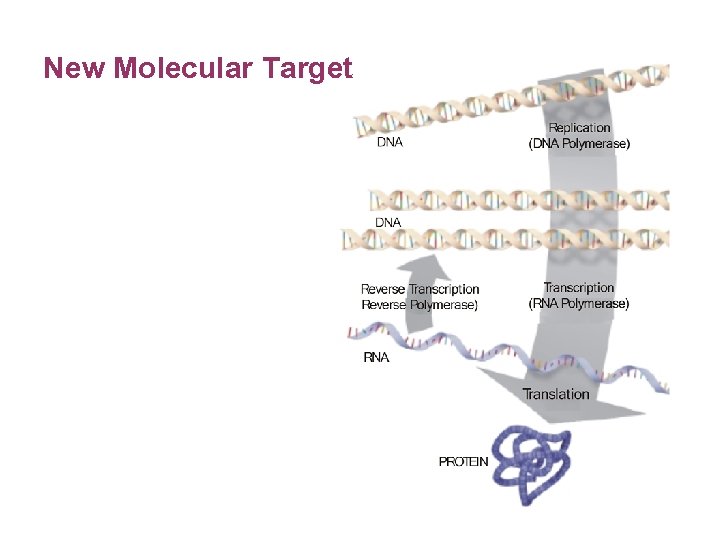 New Molecular Target
New Molecular Target
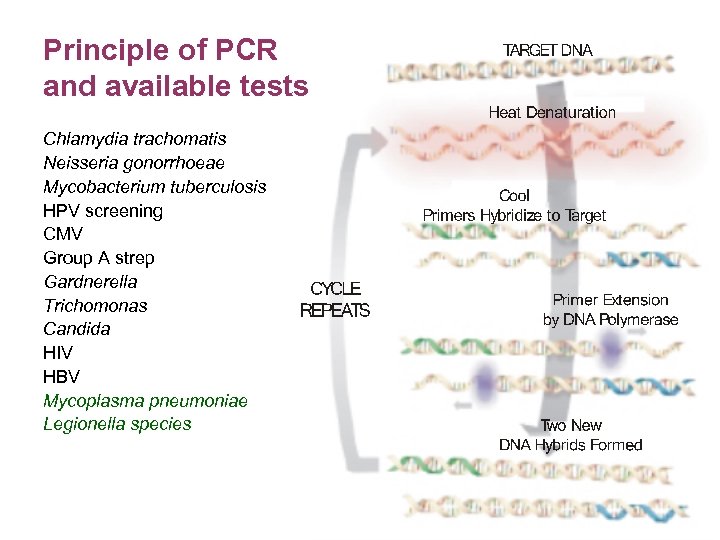 Principle of PCR and available tests Chlamydia trachomatis Neisseria gonorrhoeae Mycobacterium tuberculosis HPV screening CMV Group A strep Gardnerella Trichomonas Candida HIV HBV Mycoplasma pneumoniae Legionella species
Principle of PCR and available tests Chlamydia trachomatis Neisseria gonorrhoeae Mycobacterium tuberculosis HPV screening CMV Group A strep Gardnerella Trichomonas Candida HIV HBV Mycoplasma pneumoniae Legionella species
 Minerva Kits - Step by Step 1 - Sample Preparation sputum provoked sputum BAL nasopharyngeal swabs nasal and pharyngeal secretions
Minerva Kits - Step by Step 1 - Sample Preparation sputum provoked sputum BAL nasopharyngeal swabs nasal and pharyngeal secretions
 Minerva Kits - Step by Step 2 – DNA Extraction DNA extraction necessary for all sample materials; swabs might work without extraction all standard DNA extraction kits for total DNA preparation provide good quality templates for PCR 50 µl of DNA extract good for 10 individual reactions
Minerva Kits - Step by Step 2 – DNA Extraction DNA extraction necessary for all sample materials; swabs might work without extraction all standard DNA extraction kits for total DNA preparation provide good quality templates for PCR 50 µl of DNA extract good for 10 individual reactions
 Minerva Kits - Step by Step 3 – Reaction Setup • rehydration of the lyophylised components • master mix setup according to the handbook
Minerva Kits - Step by Step 3 – Reaction Setup • rehydration of the lyophylised components • master mix setup according to the handbook
 Minerva Kits - Step by Step 4 – Cycling • preincubation required for hotstart polymerases • primer extension at optimal temperature for high specificity, no adaptation required! • cycle number optimized for highest sensitivity • short program for modern cyclers
Minerva Kits - Step by Step 4 – Cycling • preincubation required for hotstart polymerases • primer extension at optimal temperature for high specificity, no adaptation required! • cycle number optimized for highest sensitivity • short program for modern cyclers
 Minerva Kits - Step by Step 5 – Agarose Gel Electrophoresis • separation of the PCR amplicons by charge and size in an electric field • standard agarose gel electrophoresis as established in the lab • staining of the DNA and documentation of the results according to the standard procedure in the lab total hands-on time 30 min, results within 120 to 180 min
Minerva Kits - Step by Step 5 – Agarose Gel Electrophoresis • separation of the PCR amplicons by charge and size in an electric field • standard agarose gel electrophoresis as established in the lab • staining of the DNA and documentation of the results according to the standard procedure in the lab total hands-on time 30 min, results within 120 to 180 min
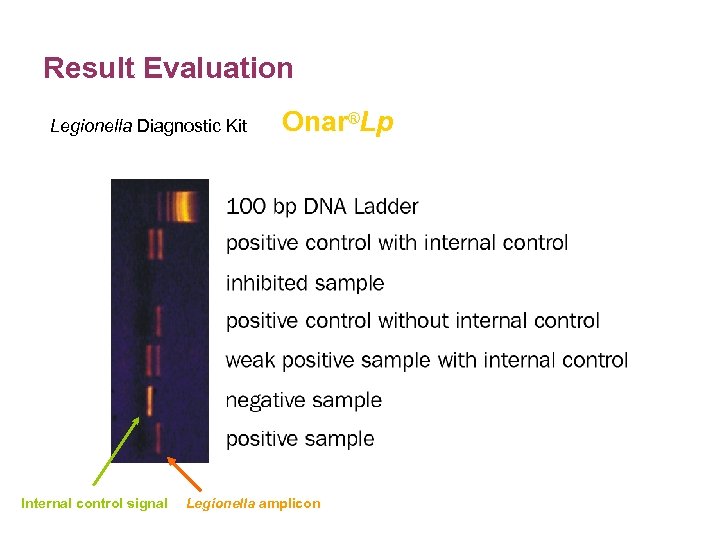 Result Evaluation Legionella Diagnostic Kit Internal control signal Onar®Lp Legionella amplicon
Result Evaluation Legionella Diagnostic Kit Internal control signal Onar®Lp Legionella amplicon
 Technical Features Venor®Mp Onar®Lp • specificity for Mycoplasma pneumoniae subtypes 1 and 2 variants 1 and 2 • detection limit 1600 particles/ml, according 8 particles/reaction • upper limit 72 µg/ml DNA, corresponds 4 x 108 particles • reproducibility 100 % • cross reactivity not found • supports UNG incorporation for carry-over-prevention • specific for Legionella pneumophila SG 1 -14 L. longbeachae SG 1 and 2 L. dumoffii, L. bozemanii L. micadadei L. jordanis, etc. • detection limit < 10 particles/reaction • reproducibility 100 % • cross reactivity not found • supports UNG incorporation for carry-over-prevention
Technical Features Venor®Mp Onar®Lp • specificity for Mycoplasma pneumoniae subtypes 1 and 2 variants 1 and 2 • detection limit 1600 particles/ml, according 8 particles/reaction • upper limit 72 µg/ml DNA, corresponds 4 x 108 particles • reproducibility 100 % • cross reactivity not found • supports UNG incorporation for carry-over-prevention • specific for Legionella pneumophila SG 1 -14 L. longbeachae SG 1 and 2 L. dumoffii, L. bozemanii L. micadadei L. jordanis, etc. • detection limit < 10 particles/reaction • reproducibility 100 % • cross reactivity not found • supports UNG incorporation for carry-over-prevention
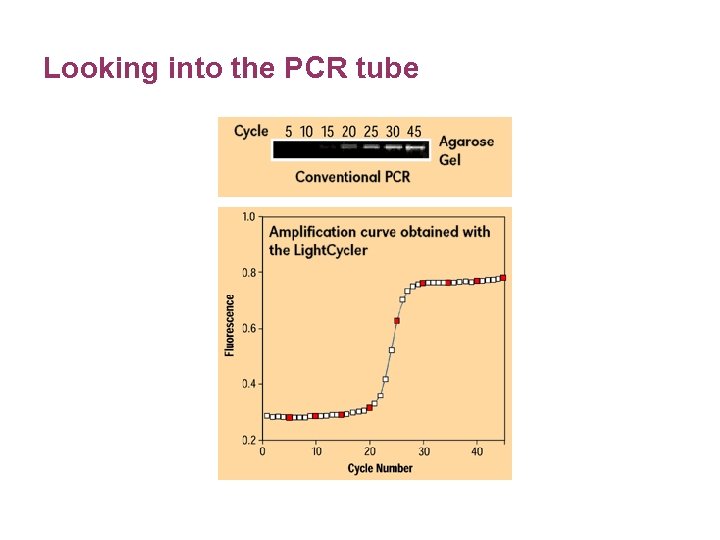 Looking into the PCR tube
Looking into the PCR tube
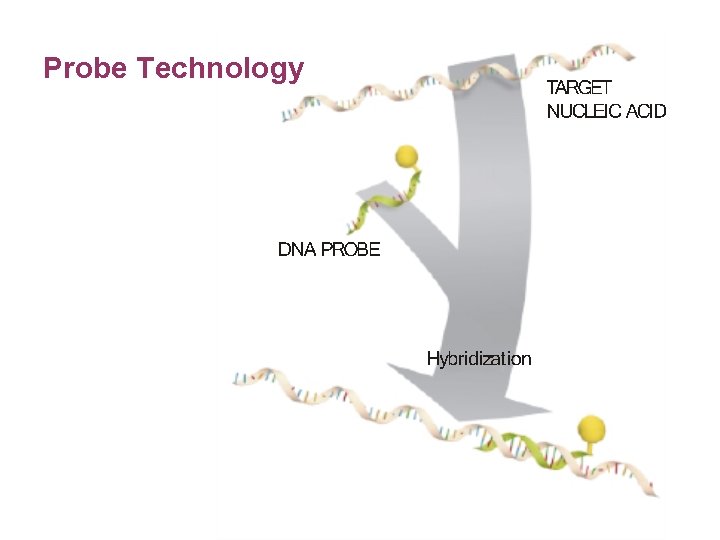 Probe Technology
Probe Technology
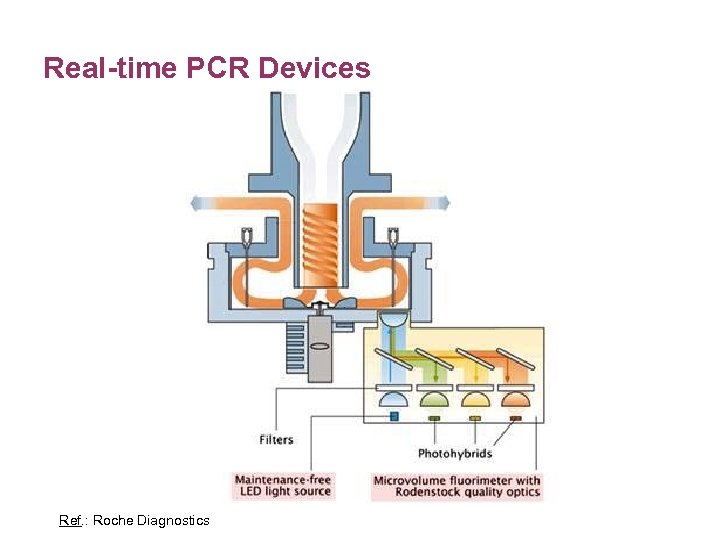 Real-time PCR Devices Ref. : Roche Diagnostics
Real-time PCR Devices Ref. : Roche Diagnostics
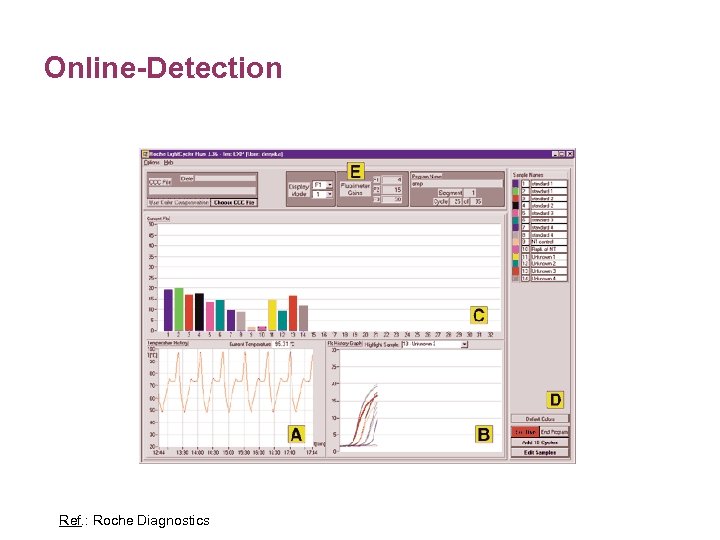 Online-Detection Ref. : Roche Diagnostics
Online-Detection Ref. : Roche Diagnostics
 Probe Technology
Probe Technology
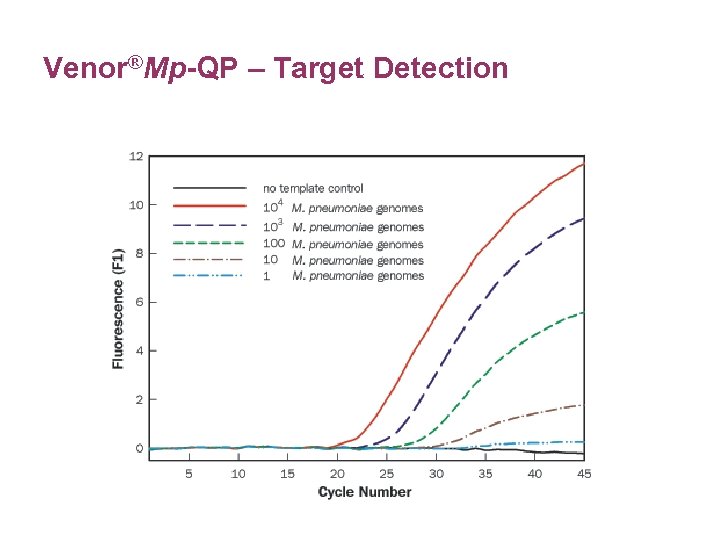 Venor®Mp-QP – Target Detection
Venor®Mp-QP – Target Detection
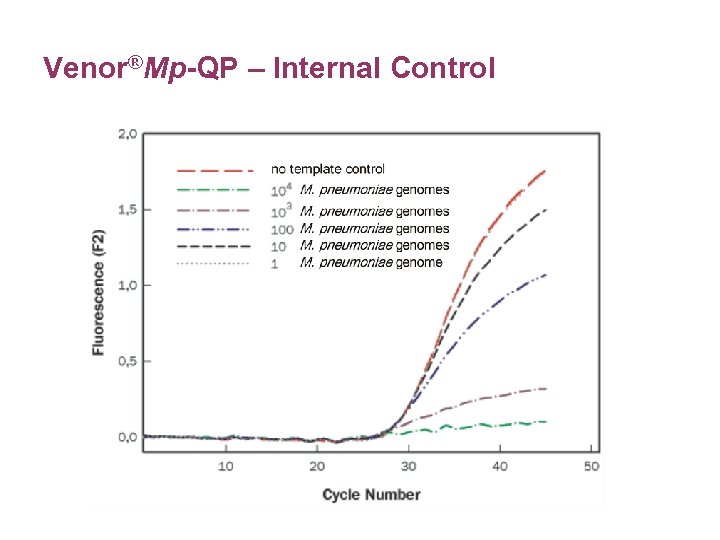 Venor®Mp-QP – Internal Control
Venor®Mp-QP – Internal Control
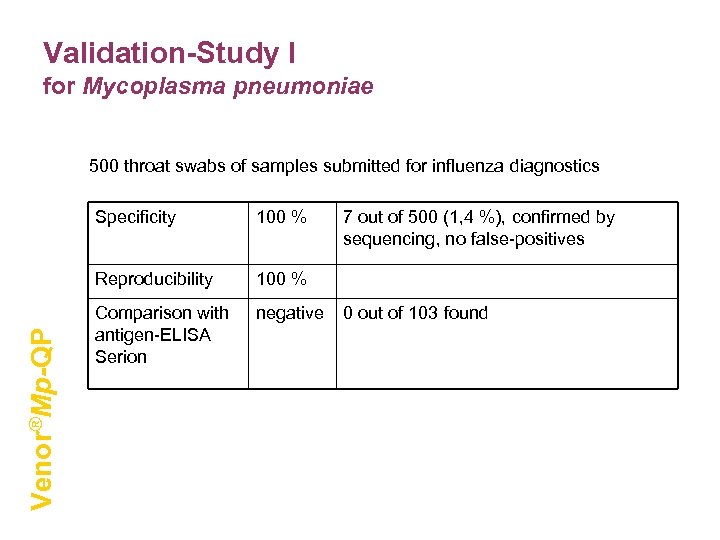 Validation-Study I for Mycoplasma pneumoniae 500 throat swabs of samples submitted for influenza diagnostics 100 % Reproducibility Venor®Mp-QP Specificity 100 % Comparison with antigen-ELISA Serion negative 7 out of 500 (1, 4 %), confirmed by sequencing, no false-positives 0 out of 103 found
Validation-Study I for Mycoplasma pneumoniae 500 throat swabs of samples submitted for influenza diagnostics 100 % Reproducibility Venor®Mp-QP Specificity 100 % Comparison with antigen-ELISA Serion negative 7 out of 500 (1, 4 %), confirmed by sequencing, no false-positives 0 out of 103 found
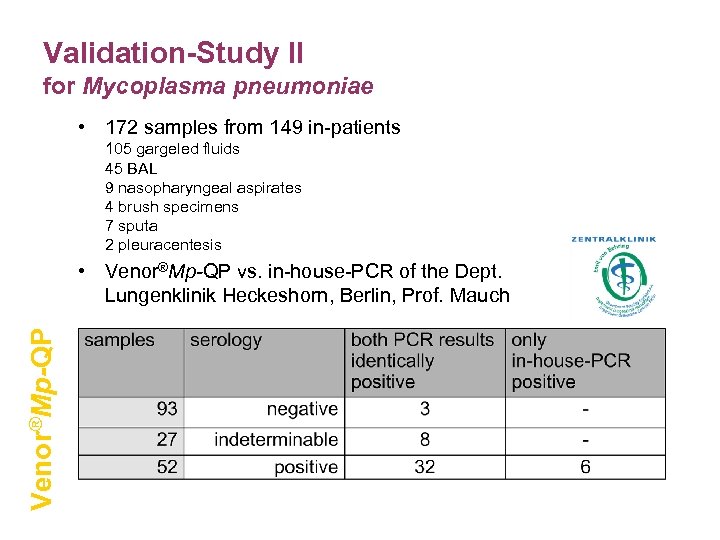 Validation-Study II for Mycoplasma pneumoniae • 172 samples from 149 in-patients 105 gargeled fluids 45 BAL 9 nasopharyngeal aspirates 4 brush specimens 7 sputa 2 pleuracentesis Venor®Mp-QP • Venor®Mp-QP vs. in-house-PCR of the Dept. Lungenklinik Heckeshorn, Berlin, Prof. Mauch
Validation-Study II for Mycoplasma pneumoniae • 172 samples from 149 in-patients 105 gargeled fluids 45 BAL 9 nasopharyngeal aspirates 4 brush specimens 7 sputa 2 pleuracentesis Venor®Mp-QP • Venor®Mp-QP vs. in-house-PCR of the Dept. Lungenklinik Heckeshorn, Berlin, Prof. Mauch
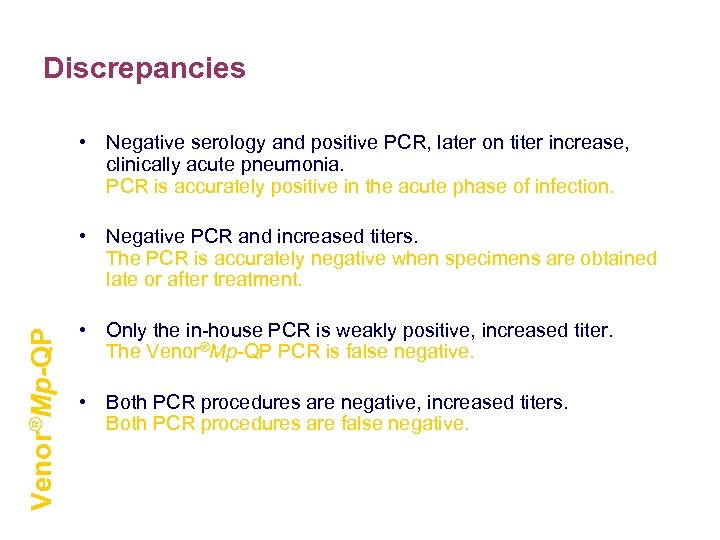 Discrepancies • Negative serology and positive PCR, later on titer increase, clinically acute pneumonia. PCR is accurately positive in the acute phase of infection. Venor®Mp-QP • Negative PCR and increased titers. The PCR is accurately negative when specimens are obtained late or after treatment. • Only the in-house PCR is weakly positive, increased titer. The Venor®Mp-QP PCR is false negative. • Both PCR procedures are negative, increased titers. Both PCR procedures are false negative.
Discrepancies • Negative serology and positive PCR, later on titer increase, clinically acute pneumonia. PCR is accurately positive in the acute phase of infection. Venor®Mp-QP • Negative PCR and increased titers. The PCR is accurately negative when specimens are obtained late or after treatment. • Only the in-house PCR is weakly positive, increased titer. The Venor®Mp-QP PCR is false negative. • Both PCR procedures are negative, increased titers. Both PCR procedures are false negative.
 Features – real-time PCR Kits • highly reliable due to specific probe system (>99, 9 % specificity) no false positive results in 850 patient samples tested • highly sensitiv: positive cut-off 10 to 100 DNA copies/reaction • reproducibility 99, 9 % • one protocoll for all respiratory targets: no single tube multiplex for multiple pathogens recommended • easy and quick handling: Results within 30 to 90 min, Hands-on-Time 20 min
Features – real-time PCR Kits • highly reliable due to specific probe system (>99, 9 % specificity) no false positive results in 850 patient samples tested • highly sensitiv: positive cut-off 10 to 100 DNA copies/reaction • reproducibility 99, 9 % • one protocoll for all respiratory targets: no single tube multiplex for multiple pathogens recommended • easy and quick handling: Results within 30 to 90 min, Hands-on-Time 20 min
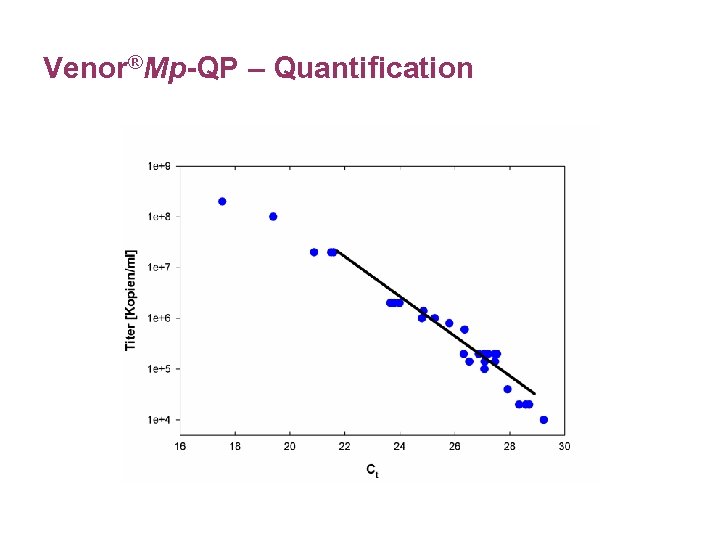 Venor®Mp-QP – Quantification
Venor®Mp-QP – Quantification
 Interpretation of Quantitative Results • Data indicates course and success of treatment. • Data provides only an idea of the general infection status, no correlation possible between the quantitative result and the health status of the patient. • The influence of the sampling techniques, time point of sampling and the type of sample material on the quantitative result need to be investigated.
Interpretation of Quantitative Results • Data indicates course and success of treatment. • Data provides only an idea of the general infection status, no correlation possible between the quantitative result and the health status of the patient. • The influence of the sampling techniques, time point of sampling and the type of sample material on the quantitative result need to be investigated.
 Special Requirements for Sampling and Sample Transfer Prefered sample material: BAL > Sputum > Swab Preparation: Refridgerated dispatch to the lab, no transfer medium For swabs only: rinsed in buffer, heat-inactivated Squeezed on transfer paper Stability no treatment: 2 - 3 day heat inactivation: >5 days transfer paper: > 8 years ! No vital particles required!
Special Requirements for Sampling and Sample Transfer Prefered sample material: BAL > Sputum > Swab Preparation: Refridgerated dispatch to the lab, no transfer medium For swabs only: rinsed in buffer, heat-inactivated Squeezed on transfer paper Stability no treatment: 2 - 3 day heat inactivation: >5 days transfer paper: > 8 years ! No vital particles required!
 Benefits of PCR in the Diagnosis of Respiratory Infection • sensitive and specific method for the detection of acute Mycoplasma and Legionella infections • especially useful for the confirmation of a clinical diagnosis in the early stage of respiratory infection • direct detection of slow growing pathogens in various sample material • shorter time-to-result: effective treatment of patient • highest performance: > 99 % specificity, > 99 % reproducibility • detection of multiple respiratory pathogens per run and sample • stable sample material, easy sampling, identical sample prep for all specimens
Benefits of PCR in the Diagnosis of Respiratory Infection • sensitive and specific method for the detection of acute Mycoplasma and Legionella infections • especially useful for the confirmation of a clinical diagnosis in the early stage of respiratory infection • direct detection of slow growing pathogens in various sample material • shorter time-to-result: effective treatment of patient • highest performance: > 99 % specificity, > 99 % reproducibility • detection of multiple respiratory pathogens per run and sample • stable sample material, easy sampling, identical sample prep for all specimens
 General benefits of real-time PCR • quantitative determination of infection level • results are available immediately after PCR in an electronic form (GLP conformity) • highly practicable: – highest speed (1 hour testing time) – high-throughput automation possible (up to 15. 000 samples/day) • amplification and detection are combined in a single step and in a single closed tube: lowest risk of cross contamination • no need for time-consuming numerous post-PCR manual steps • hybridization for result confirmation and highest specificity included
General benefits of real-time PCR • quantitative determination of infection level • results are available immediately after PCR in an electronic form (GLP conformity) • highly practicable: – highest speed (1 hour testing time) – high-throughput automation possible (up to 15. 000 samples/day) • amplification and detection are combined in a single step and in a single closed tube: lowest risk of cross contamination • no need for time-consuming numerous post-PCR manual steps • hybridization for result confirmation and highest specificity included
 Presentation available through: www. minerva-biolabs. com Please click on the headline in the news ticker.
Presentation available through: www. minerva-biolabs. com Please click on the headline in the news ticker.


