b8461205bbceb46ad287968d4e818209.ppt
- Количество слайдов: 14
 Temporal Reasoning and Planning in Medicine Temporal Reasoning In Medical Information Systems (I) Yuval Shahar M. D. , Ph. D.
Temporal Reasoning and Planning in Medicine Temporal Reasoning In Medical Information Systems (I) Yuval Shahar M. D. , Ph. D.
 Time as Symbolic Tokens • “Chest pain, substernal, lasting less than 20 minutes” (the INTERNIST-I system for internal-medicine diagnosis [Miller et al. , 1982]) – No ability to note [at least a potential] contradiction with “Chest pain, substernal, lasting more than 20 minutes” • “Significant organisms isolated within the past 30 days” (The MYCIN system for infectious-diseases diagnosis and therapy) • Answers are always Yes or No (Boolean predicates) and no further reasoning on the structure of the token is possible • Typically, no transaction or valid times, only a duration
Time as Symbolic Tokens • “Chest pain, substernal, lasting less than 20 minutes” (the INTERNIST-I system for internal-medicine diagnosis [Miller et al. , 1982]) – No ability to note [at least a potential] contradiction with “Chest pain, substernal, lasting more than 20 minutes” • “Significant organisms isolated within the past 30 days” (The MYCIN system for infectious-diseases diagnosis and therapy) • Answers are always Yes or No (Boolean predicates) and no further reasoning on the structure of the token is possible • Typically, no transaction or valid times, only a duration
 Fagan’s Ventilator Management (VM) System (Fagan, 1980) • Designed for management of patients on ventilators in ICUs • Rule based; rules were specific to contexts (states) • Explicit reasoning about time units, rates of change • Context-specific classifications mapped parameters into a context-sensitive range of values (e. g. , acceptable), enabling context-free rules and context-free aggregation of abstractions • Expiration dates of parameters represented in a good-for slot => constants, continuous, volunteered, deduced parameter types • Special state-change rules inferred a new context (mode) • Data could not be accepted out of temporal order (no valid time)
Fagan’s Ventilator Management (VM) System (Fagan, 1980) • Designed for management of patients on ventilators in ICUs • Rule based; rules were specific to contexts (states) • Explicit reasoning about time units, rates of change • Context-specific classifications mapped parameters into a context-sensitive range of values (e. g. , acceptable), enabling context-free rules and context-free aggregation of abstractions • Expiration dates of parameters represented in a good-for slot => constants, continuous, volunteered, deduced parameter types • Special state-change rules inferred a new context (mode) • Data could not be accepted out of temporal order (no valid time)
 Blum’s Rx System (1982) • Analyzed time-oriented clinical databases to produce a set of possible causal relations • Used two modules in succession: – A Discovery Module, for automated discovery of statistical relations – A Study Module, to rule out spurious correlations, by using a medical knowledge base and by creating and testing a statistical model of the hypothesis • Applied to data in the Stanford American Rheumatic Association Medical Information System (ARAMIS), which evolved from the Time Oriented Database (TOD) • ARAMIS and TOD are three-dimensional historical databases – a patient ID indexes
Blum’s Rx System (1982) • Analyzed time-oriented clinical databases to produce a set of possible causal relations • Used two modules in succession: – A Discovery Module, for automated discovery of statistical relations – A Study Module, to rule out spurious correlations, by using a medical knowledge base and by creating and testing a statistical model of the hypothesis • Applied to data in the Stanford American Rheumatic Association Medical Information System (ARAMIS), which evolved from the Time Oriented Database (TOD) • ARAMIS and TOD are three-dimensional historical databases – a patient ID indexes
 Data Representation in Rx • Point Events (e. g. , laboratory test) • Interval Events (e. g. , a disease spanning several visits) – required an extension to the TOD • Used a hierarchical derivation tree: Event A can be defined in terms of events B 1, B 2; these can be defined by C 11, C 12, and C 21, C 22… – to assess a value, traverse its derivation tree and collect all values • Sometimes a clinical parameter needs to be assessed when not measured (a latent variable), using proxy variables • Time-dependent database access functions (delayed_action, delayed_effect, previous_value) • Parameter-specific knowledge determined inter-episode gaps
Data Representation in Rx • Point Events (e. g. , laboratory test) • Interval Events (e. g. , a disease spanning several visits) – required an extension to the TOD • Used a hierarchical derivation tree: Event A can be defined in terms of events B 1, B 2; these can be defined by C 11, C 12, and C 21, C 22… – to assess a value, traverse its derivation tree and collect all values • Sometimes a clinical parameter needs to be assessed when not measured (a latent variable), using proxy variables • Time-dependent database access functions (delayed_action, delayed_effect, previous_value) • Parameter-specific knowledge determined inter-episode gaps
 Downs’ Medical-record Summarization Program (1986) • Designed specifically to automate the summarization of online medical records • Like Rx, used the Stanford ARAMIS database • A knowledge base represented two classes of parameters: – abnormal attributes (abnormal findings) – derived attributes (including diseases that can be inferred from data) • Each abnormal attribute points to a list of derived attributes that should be considered if its value is True • Inference based on a hypothetico-deductive approach: Data-driven hypothesis generation produces a list of potential diagnoses; discrimination among competing hypotheses follows, using positive and negative evidence • Probabilistic, Bayesian reasoning: Prior likelihood ratios are updated by each relevant datum • Temporal predicates used, such as “the last 5 creatinine values are above 2. 0”
Downs’ Medical-record Summarization Program (1986) • Designed specifically to automate the summarization of online medical records • Like Rx, used the Stanford ARAMIS database • A knowledge base represented two classes of parameters: – abnormal attributes (abnormal findings) – derived attributes (including diseases that can be inferred from data) • Each abnormal attribute points to a list of derived attributes that should be considered if its value is True • Inference based on a hypothetico-deductive approach: Data-driven hypothesis generation produces a list of potential diagnoses; discrimination among competing hypotheses follows, using positive and negative evidence • Probabilistic, Bayesian reasoning: Prior likelihood ratios are updated by each relevant datum • Temporal predicates used, such as “the last 5 creatinine values are above 2. 0”
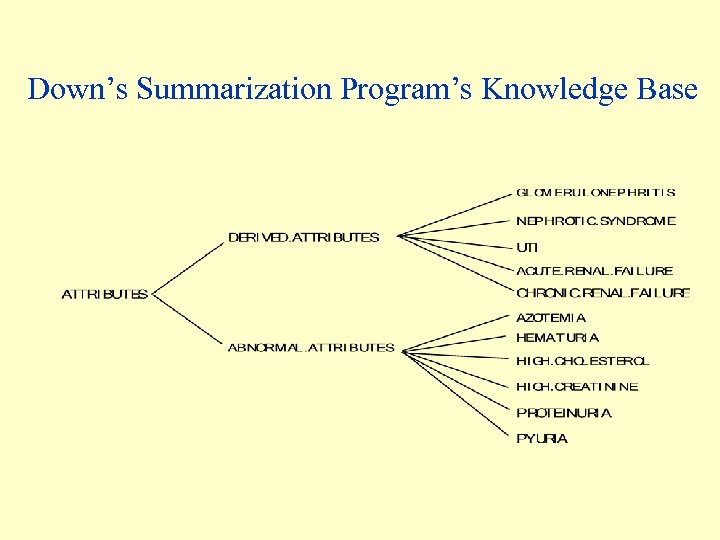 Down’s Summarization Program’s Knowledge Base
Down’s Summarization Program’s Knowledge Base
 De Zegher-Geets’ IDEFIX System (1987) • Goal: Intelligent summarization of online patient records – Explain, for a given visit, all manifestations in that visit, given previous data • Influenced by and extends Down’s summarization program • Use the ARAMIS database, especially for systemic lupus erythematosus (SLE) patients • Updated disease likelihoods by Bayesian odds-update functions • Distinguished static medical knowledge from dynamic patient data • Three levels of abstraction in the knowledge base’s ontology: – abnormal primary attributes (APAs) such as presence of protein in urine – abnormal states, such as nephrotic syndrome – diseases, such as SLE-related nephritis
De Zegher-Geets’ IDEFIX System (1987) • Goal: Intelligent summarization of online patient records – Explain, for a given visit, all manifestations in that visit, given previous data • Influenced by and extends Down’s summarization program • Use the ARAMIS database, especially for systemic lupus erythematosus (SLE) patients • Updated disease likelihoods by Bayesian odds-update functions • Distinguished static medical knowledge from dynamic patient data • Three levels of abstraction in the knowledge base’s ontology: – abnormal primary attributes (APAs) such as presence of protein in urine – abnormal states, such as nephrotic syndrome – diseases, such as SLE-related nephritis
 The IDEFIX Inference Methods • Two phase inference: – Goal-driven strategy explains given APAs, using a list of complications of the current disease – Followed by a data-driven strategy that tries to explain remaining APAs using a Cover and Differentiate approach based on oddslikelihood ratios, similar to Downs’ program • Unlike Downs’ program, used severity levels using severity fiunctions for manifestations as well as for states or diseases • Time-Oriented Probabilistic Functions (TOPFs) returned conditional probability of disease D given manifestation M as a function of a time interval (e. g. , duration of D) – Used only to compute positive evidence; did not use context, severity
The IDEFIX Inference Methods • Two phase inference: – Goal-driven strategy explains given APAs, using a list of complications of the current disease – Followed by a data-driven strategy that tries to explain remaining APAs using a Cover and Differentiate approach based on oddslikelihood ratios, similar to Downs’ program • Unlike Downs’ program, used severity levels using severity fiunctions for manifestations as well as for states or diseases • Time-Oriented Probabilistic Functions (TOPFs) returned conditional probability of disease D given manifestation M as a function of a time interval (e. g. , duration of D) – Used only to compute positive evidence; did not use context, severity
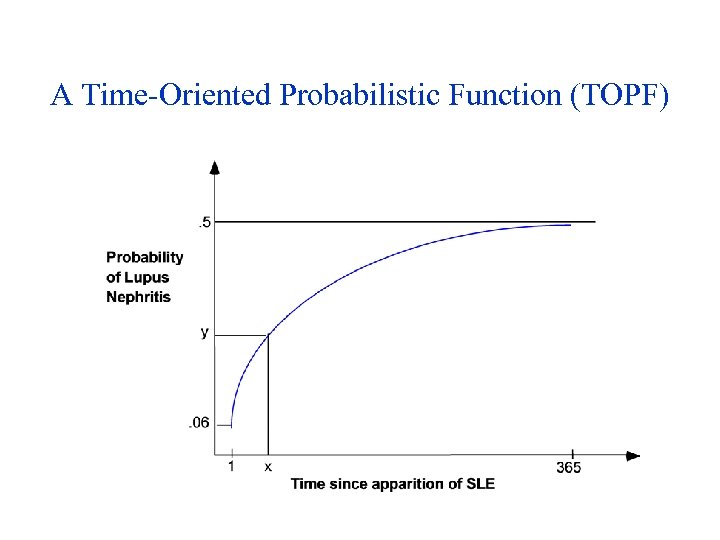 A Time-Oriented Probabilistic Function (TOPF)
A Time-Oriented Probabilistic Function (TOPF)
![Russ's Temporal Control Structure [TCS] System (1983 -1991) • Manages data dependencies over time Russ's Temporal Control Structure [TCS] System (1983 -1991) • Manages data dependencies over time](https://present5.com/presentation/b8461205bbceb46ad287968d4e818209/image-11.jpg) Russ's Temporal Control Structure [TCS] System (1983 -1991) • Manages data dependencies over time • Data driven • Supports decomposition of reasoning into static and dynamic parts • Performs the necessary bookkeeping needed to ensure propagation of information in the system as well as completeness in processing • A point-based approach with exact dating • Performs procedural abstractions • Supports hindsight as more data accumulate • Evaluated as part of a diabetic ketoacidosis monitoring system
Russ's Temporal Control Structure [TCS] System (1983 -1991) • Manages data dependencies over time • Data driven • Supports decomposition of reasoning into static and dynamic parts • Performs the necessary bookkeeping needed to ensure propagation of information in the system as well as completeness in processing • A point-based approach with exact dating • Performs procedural abstractions • Supports hindsight as more data accumulate • Evaluated as part of a diabetic ketoacidosis monitoring system
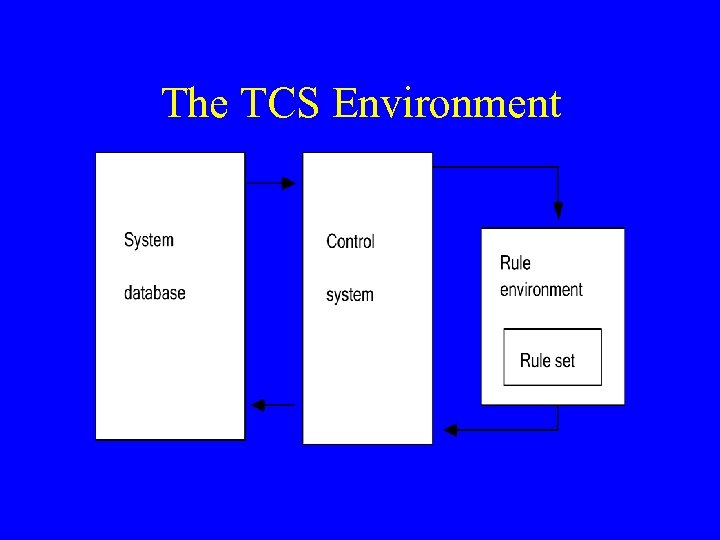 The TCS Environment
The TCS Environment
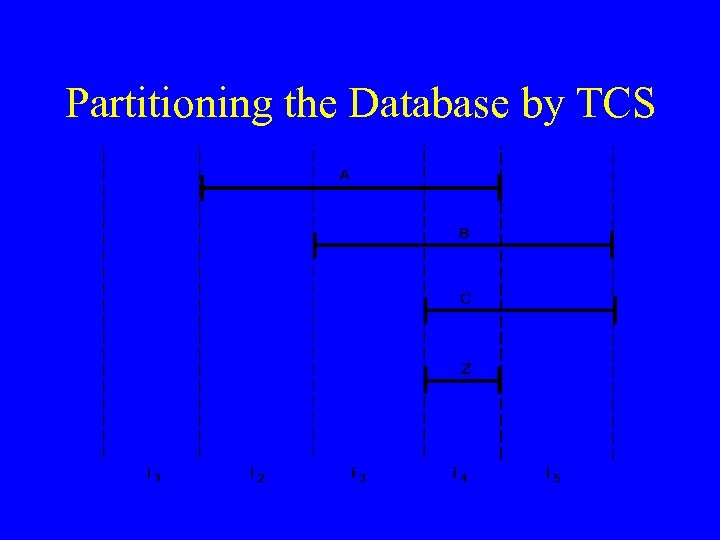 Partitioning the Database by TCS
Partitioning the Database by TCS
 A Process-Chain in TCS
A Process-Chain in TCS


