e603a3bba8b2d711b89d9da308ac4e26.ppt
- Количество слайдов: 45
 “Targeting Thymidylate Synthase in Cancer Therapy” F. G. Berger Department of Biological Sciences Center for Colon Cancer Research University of South Carolina
“Targeting Thymidylate Synthase in Cancer Therapy” F. G. Berger Department of Biological Sciences Center for Colon Cancer Research University of South Carolina
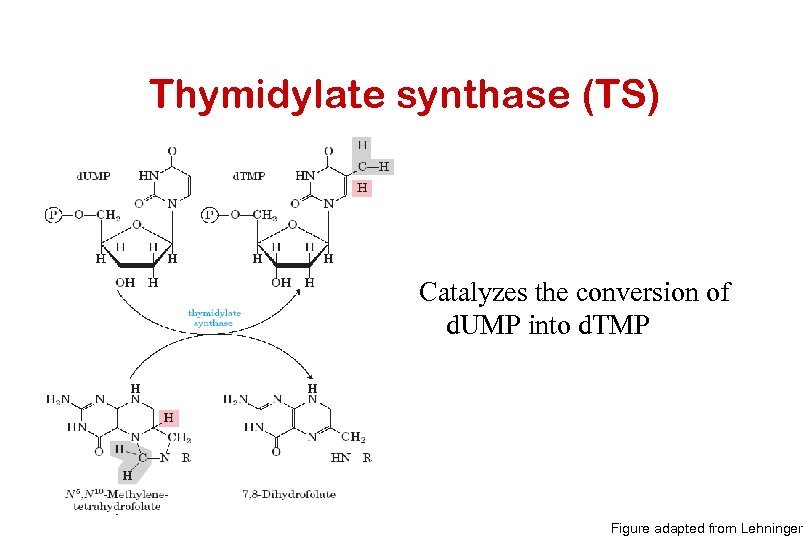 Thymidylate synthase (TS) Catalyzes the conversion of d. UMP into d. TMP Figure adapted from Lehninger
Thymidylate synthase (TS) Catalyzes the conversion of d. UMP into d. TMP Figure adapted from Lehninger
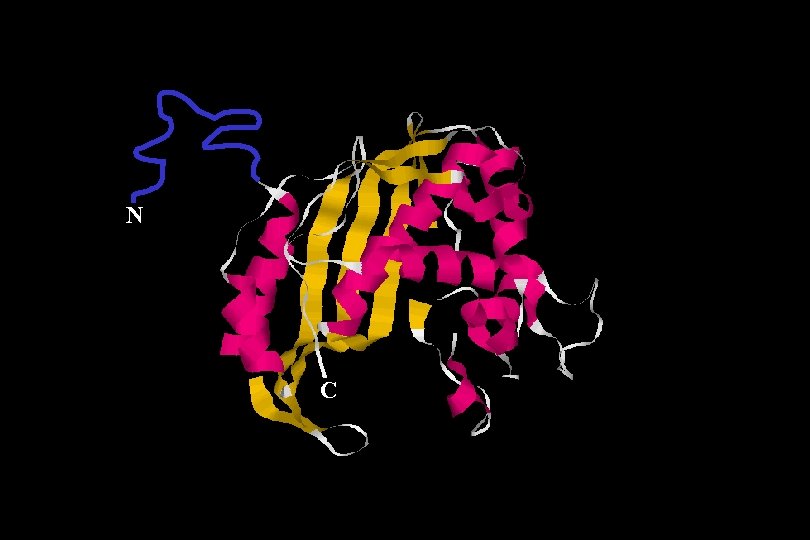 N C
N C
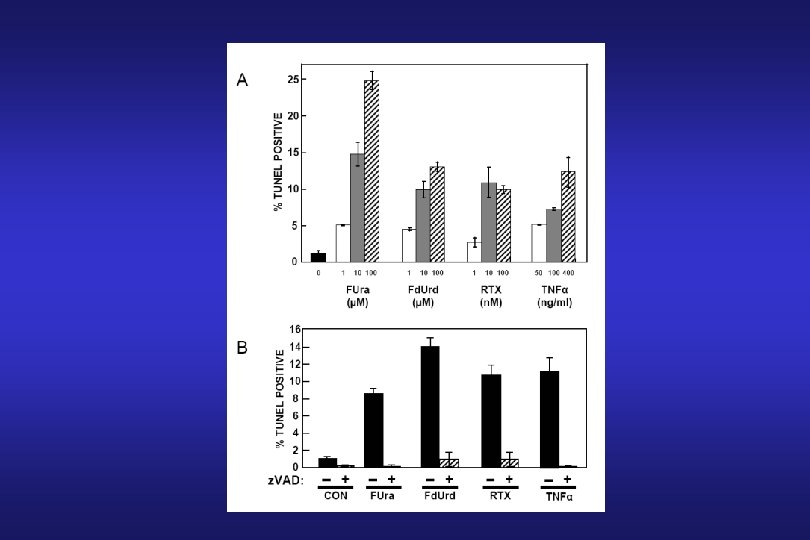
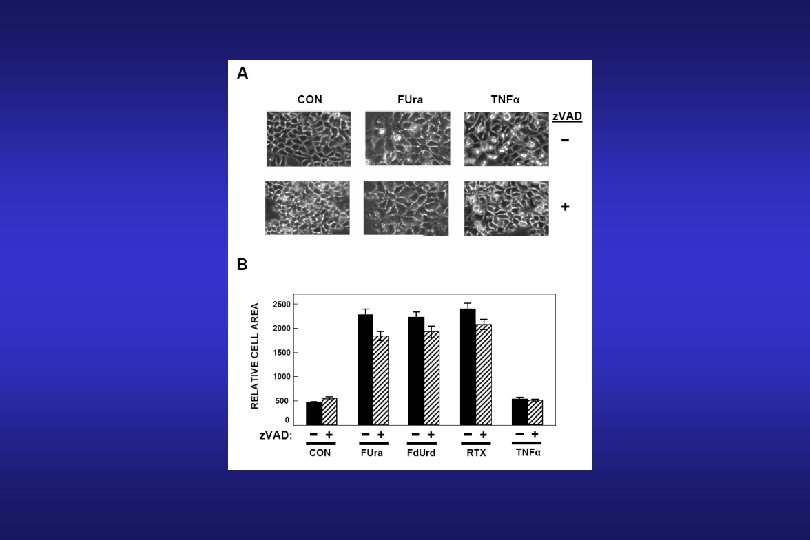
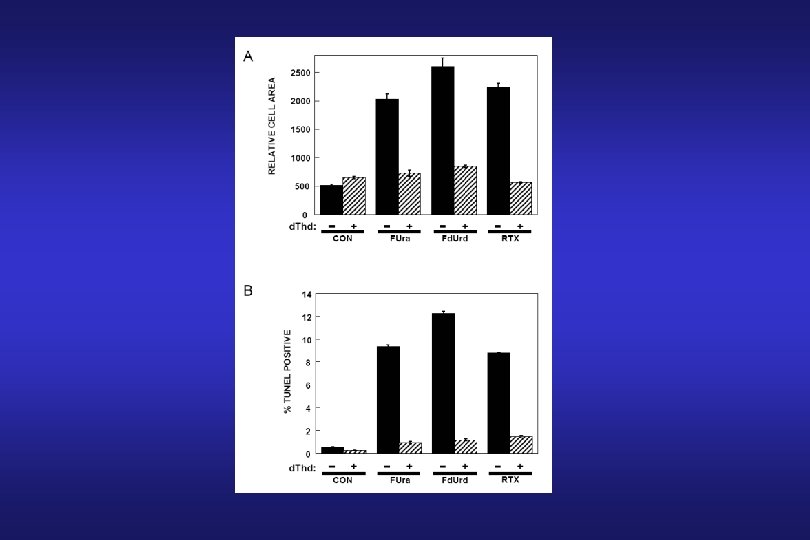
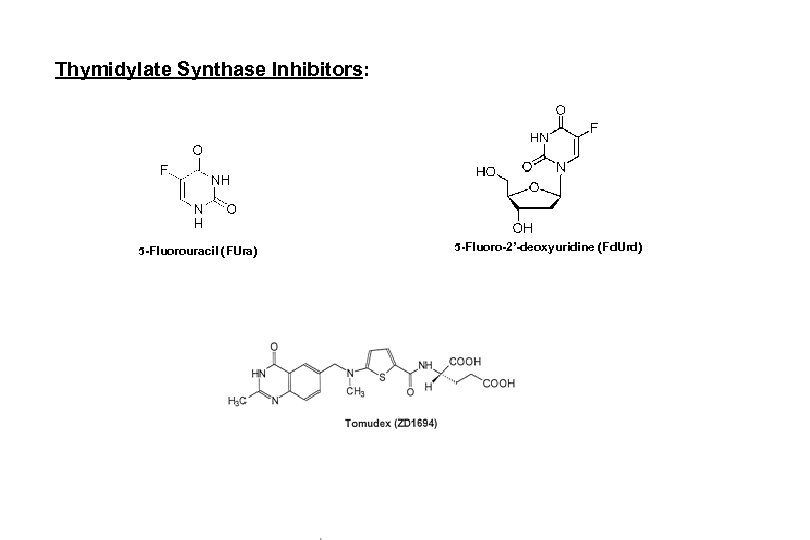 Thymidylate Synthase Inhibitors: 5 -Fluorouracil (FUra) 5 -Fluoro-2’-deoxyuridine (Fd. Urd)
Thymidylate Synthase Inhibitors: 5 -Fluorouracil (FUra) 5 -Fluoro-2’-deoxyuridine (Fd. Urd)
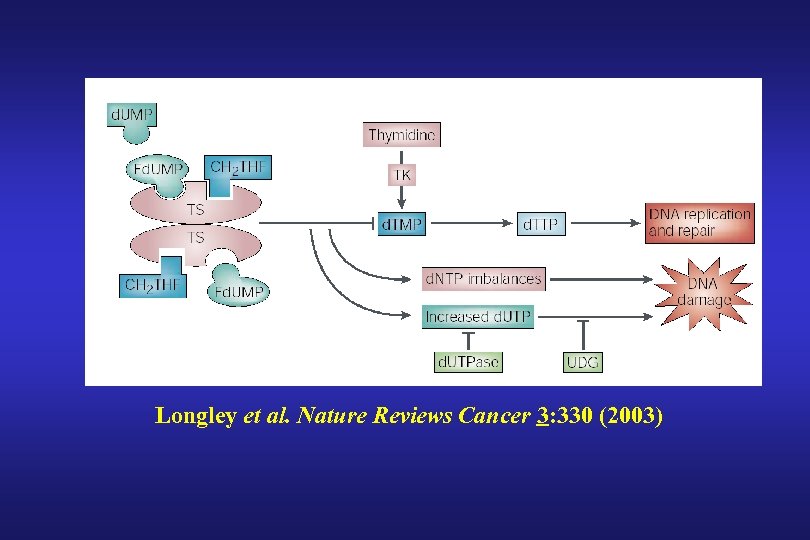 Longley et al. Nature Reviews Cancer 3: 330 (2003)
Longley et al. Nature Reviews Cancer 3: 330 (2003)
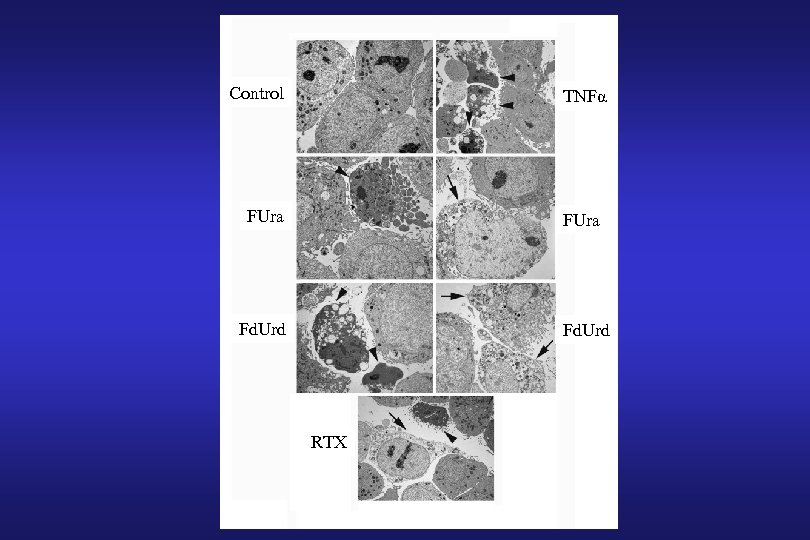 Control TNFα FUra Fd. Urd RTX
Control TNFα FUra Fd. Urd RTX
 Role of post-translational processes in TS function: ● Degradation of the TS polypeptide ● SUMO modification of TS ● Nuclear localization
Role of post-translational processes in TS function: ● Degradation of the TS polypeptide ● SUMO modification of TS ● Nuclear localization
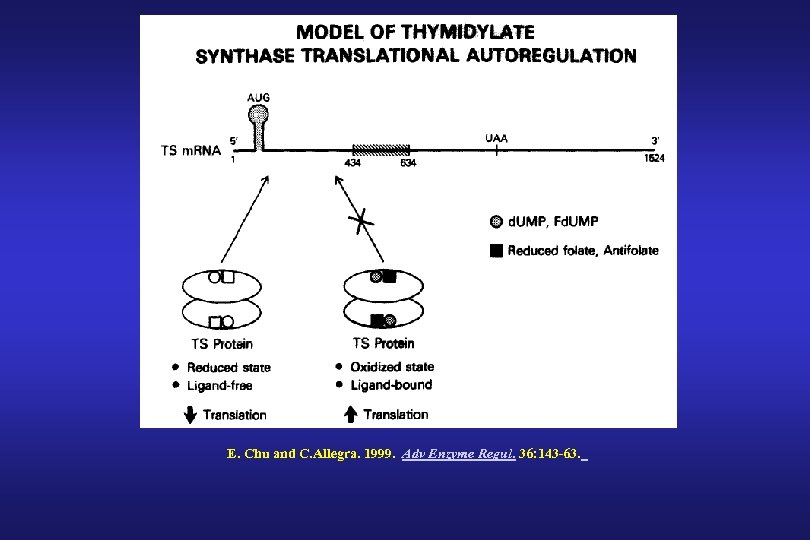 E. Chu and C. Allegra. 1999. Adv Enzyme Regul. 36: 143 -63.
E. Chu and C. Allegra. 1999. Adv Enzyme Regul. 36: 143 -63.
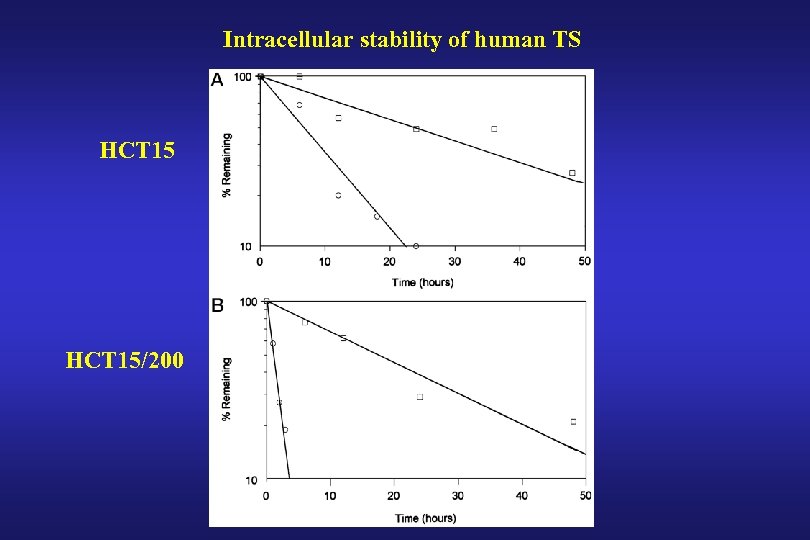 Intracellular stability of human TS HCT 15/200
Intracellular stability of human TS HCT 15/200
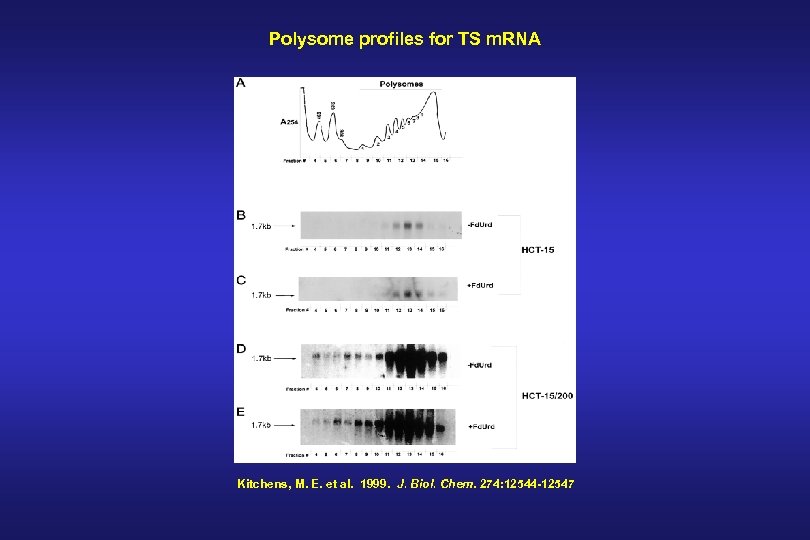 Polysome profiles for TS m. RNA Kitchens, M. E. et al. 1999. J. Biol. Chem. 274: 12544 -12547
Polysome profiles for TS m. RNA Kitchens, M. E. et al. 1999. J. Biol. Chem. 274: 12544 -12547
 ● TS is degraded intracellularly by the 26 S proteasome. ● TS is not ubiquitinylated. ● Abrogation of the ubiquitin ligation pathway does not alter TS degradation. ● Conversion of all Lys residues to Arg does not stabilize the TS polypeptide. Therefore, the TS polypeptide is degraded in a ubiquitin-independent manner.
● TS is degraded intracellularly by the 26 S proteasome. ● TS is not ubiquitinylated. ● Abrogation of the ubiquitin ligation pathway does not alter TS degradation. ● Conversion of all Lys residues to Arg does not stabilize the TS polypeptide. Therefore, the TS polypeptide is degraded in a ubiquitin-independent manner.
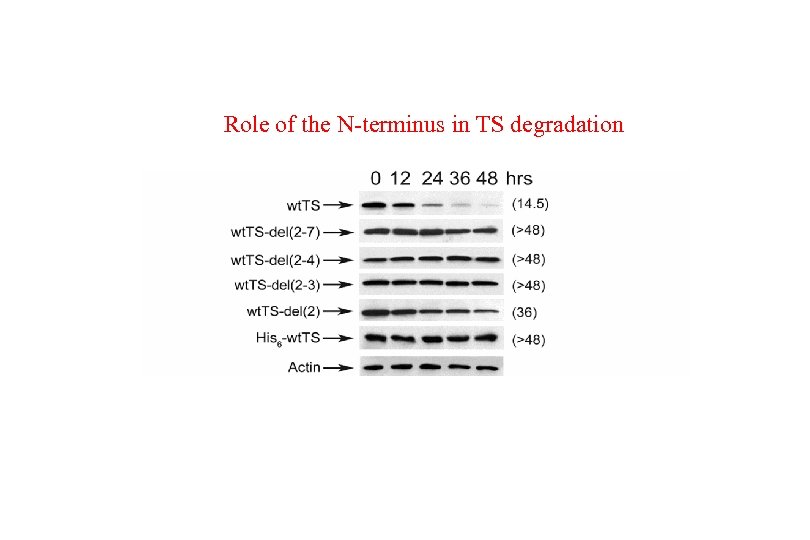 Role of the N-terminus in TS degradation
Role of the N-terminus in TS degradation
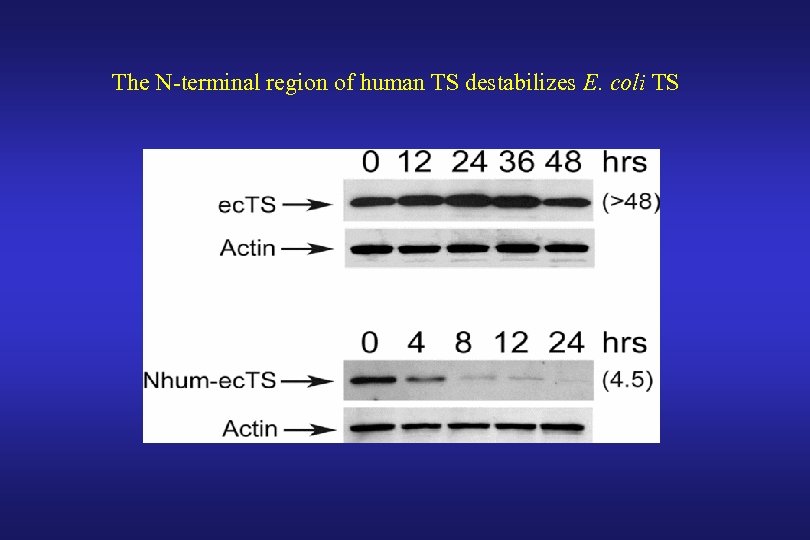 The N-terminal region of human TS destabilizes E. coli TS
The N-terminal region of human TS destabilizes E. coli TS
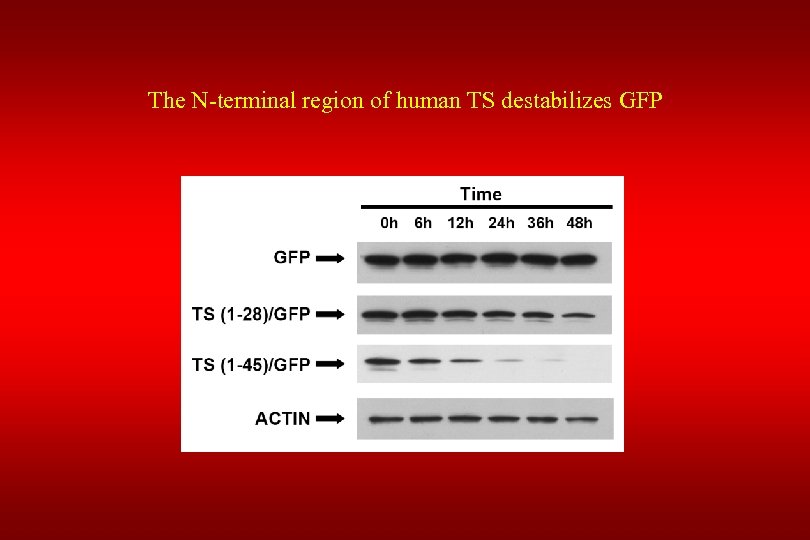 The N-terminal region of human TS destabilizes GFP
The N-terminal region of human TS destabilizes GFP
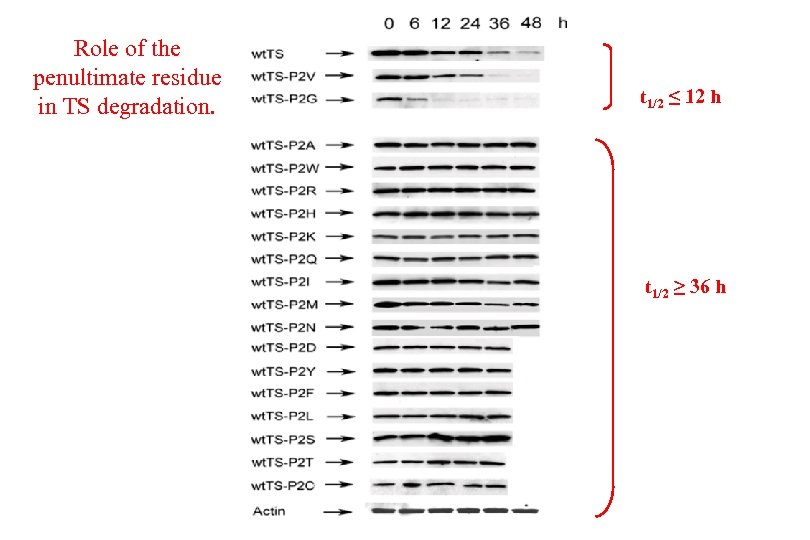 Role of the penultimate residue in TS degradation. t 1/2 ≤ 12 h t 1/2 ≥ 36 h
Role of the penultimate residue in TS degradation. t 1/2 ≤ 12 h t 1/2 ≥ 36 h
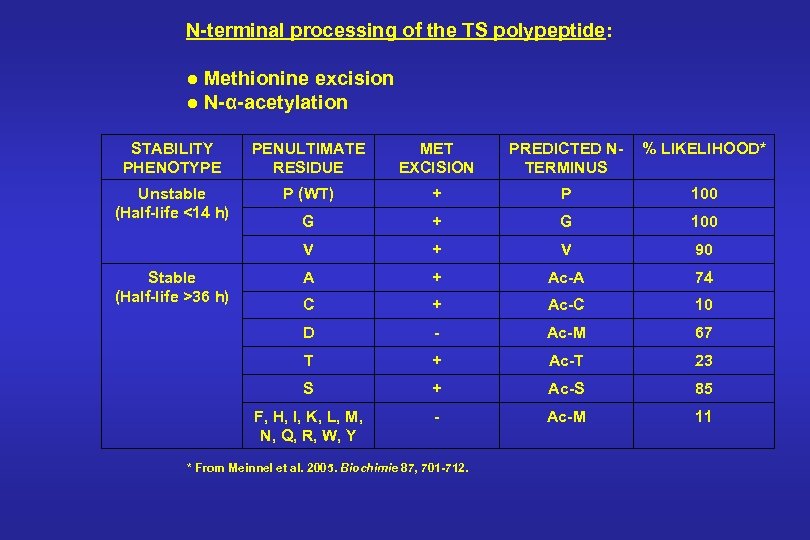 N-terminal processing of the TS polypeptide: ● Methionine excision ● N-α-acetylation STABILITY PHENOTYPE PENULTIMATE RESIDUE MET EXCISION PREDICTED NTERMINUS % LIKELIHOOD* Unstable (Half-life <14 h) P (WT) + P 100 G + G 100 V + V 90 A + Ac-A 74 C + Ac-C 10 D - Ac-M 67 T + Ac-T 23 S + Ac-S 85 F, H, I, K, L, M, N, Q, R, W, Y - Ac-M 11 Stable (Half-life >36 h) * From Meinnel et al. 2005. Biochimie 87, 701 -712.
N-terminal processing of the TS polypeptide: ● Methionine excision ● N-α-acetylation STABILITY PHENOTYPE PENULTIMATE RESIDUE MET EXCISION PREDICTED NTERMINUS % LIKELIHOOD* Unstable (Half-life <14 h) P (WT) + P 100 G + G 100 V + V 90 A + Ac-A 74 C + Ac-C 10 D - Ac-M 67 T + Ac-T 23 S + Ac-S 85 F, H, I, K, L, M, N, Q, R, W, Y - Ac-M 11 Stable (Half-life >36 h) * From Meinnel et al. 2005. Biochimie 87, 701 -712.
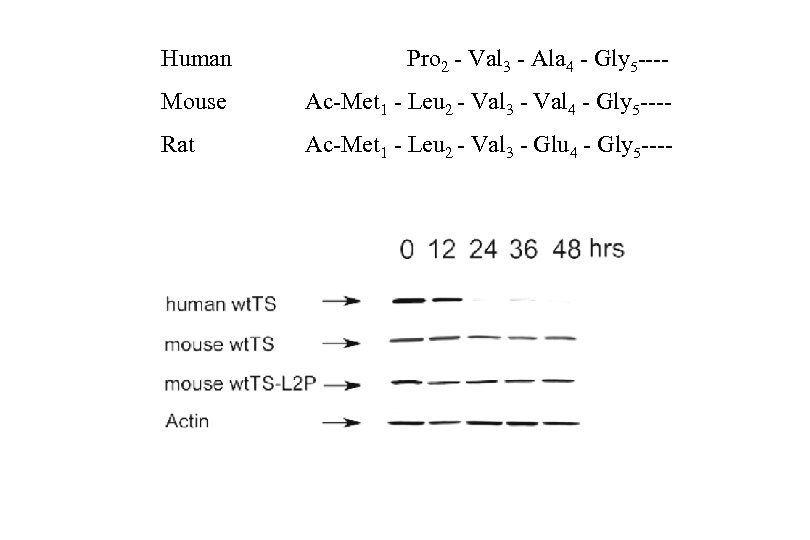 Human Pro 2 - Val 3 - Ala 4 - Gly 5 ---- Mouse Ac-Met 1 - Leu 2 - Val 3 - Val 4 - Gly 5 ---- Rat Ac-Met 1 - Leu 2 - Val 3 - Glu 4 - Gly 5 ----
Human Pro 2 - Val 3 - Ala 4 - Gly 5 ---- Mouse Ac-Met 1 - Leu 2 - Val 3 - Val 4 - Gly 5 ---- Rat Ac-Met 1 - Leu 2 - Val 3 - Glu 4 - Gly 5 ----
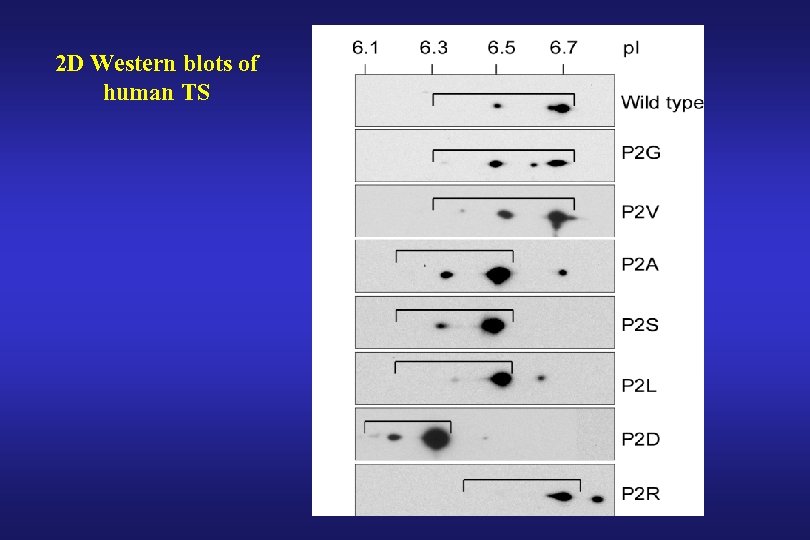 2 D Western blots of human TS
2 D Western blots of human TS
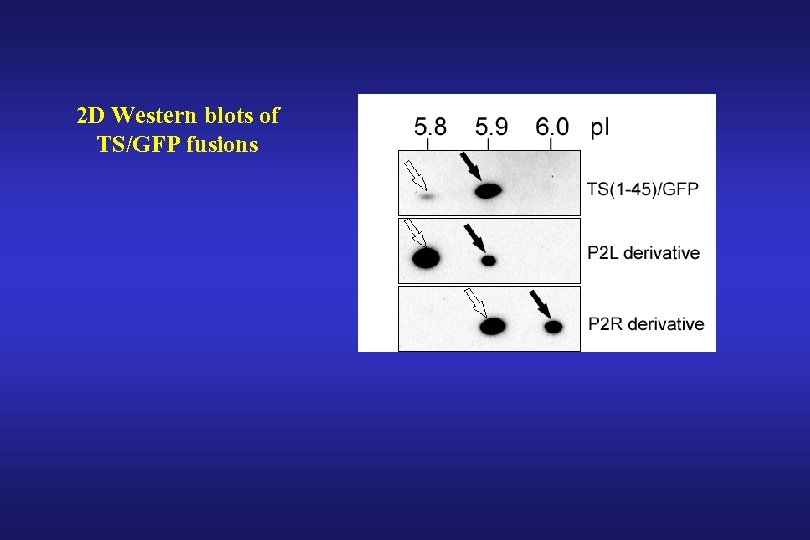 2 D Western blots of TS/GFP fusions
2 D Western blots of TS/GFP fusions
 10 PVAGSELPR 2
10 PVAGSELPR 2
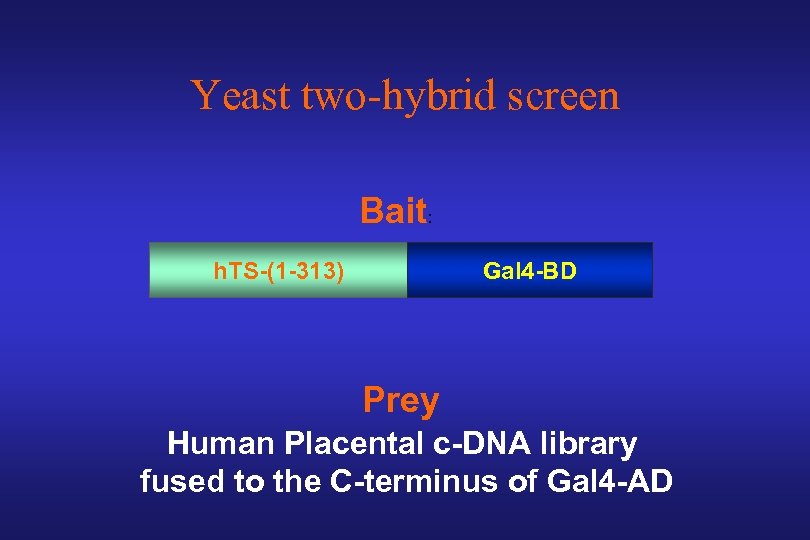 Yeast two-hybrid screen Bait: h. TS-(1 -313) Gal 4 -BD Prey Human Placental c-DNA library fused to the C-terminus of Gal 4 -AD
Yeast two-hybrid screen Bait: h. TS-(1 -313) Gal 4 -BD Prey Human Placental c-DNA library fused to the C-terminus of Gal 4 -AD
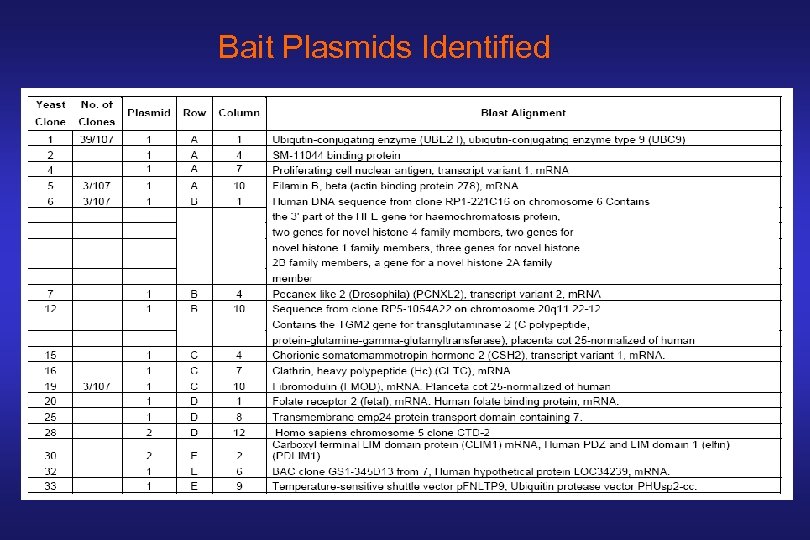 Bait Plasmids Identified
Bait Plasmids Identified
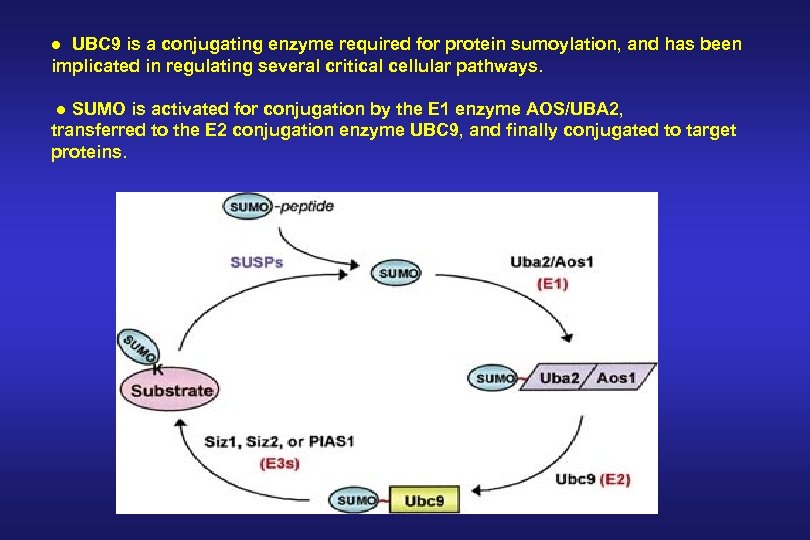 ● UBC 9 is a conjugating enzyme required for protein sumoylation, and has been implicated in regulating several critical cellular pathways. ● SUMO is activated for conjugation by the E 1 enzyme AOS/UBA 2, transferred to the E 2 conjugation enzyme UBC 9, and finally conjugated to target proteins.
● UBC 9 is a conjugating enzyme required for protein sumoylation, and has been implicated in regulating several critical cellular pathways. ● SUMO is activated for conjugation by the E 1 enzyme AOS/UBA 2, transferred to the E 2 conjugation enzyme UBC 9, and finally conjugated to target proteins.
 ● Known target proteins of sumoylation include p 53, MDM 2, PML, Ran. GAP 1, IκB, androgen receptor, and c-Jun. ● Modification of these proteins by sumoylation changes their subcellular localization, function, and/or stability. ● Sumoylation is reversible, and there at least seven mammalian SUMOspecific proteases, which are designated the SENP family proteins.
● Known target proteins of sumoylation include p 53, MDM 2, PML, Ran. GAP 1, IκB, androgen receptor, and c-Jun. ● Modification of these proteins by sumoylation changes their subcellular localization, function, and/or stability. ● Sumoylation is reversible, and there at least seven mammalian SUMOspecific proteases, which are designated the SENP family proteins.
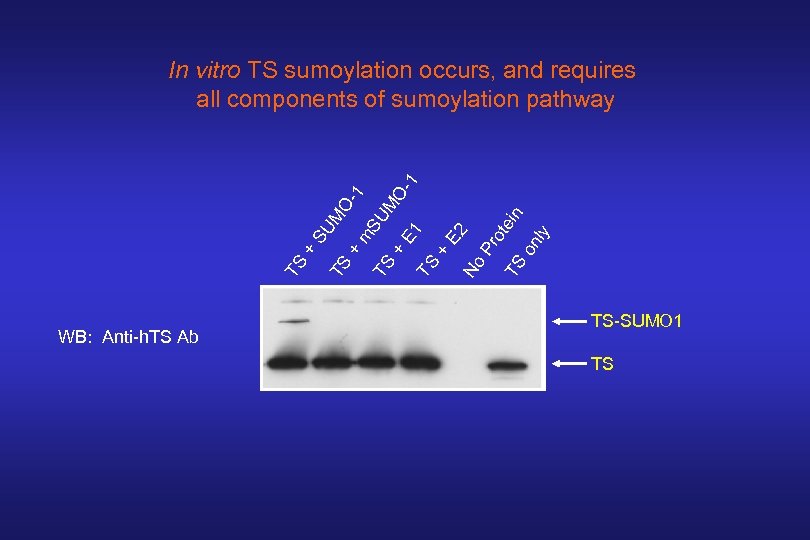 TS + SU M TS O+ 1 m SU TS M + OE 1 1 TS + E 2 No Pr o TS tein on ly In vitro TS sumoylation occurs, and requires all components of sumoylation pathway WB: Anti-h. TS Ab TS-SUMO 1 TS
TS + SU M TS O+ 1 m SU TS M + OE 1 1 TS + E 2 No Pr o TS tein on ly In vitro TS sumoylation occurs, and requires all components of sumoylation pathway WB: Anti-h. TS Ab TS-SUMO 1 TS
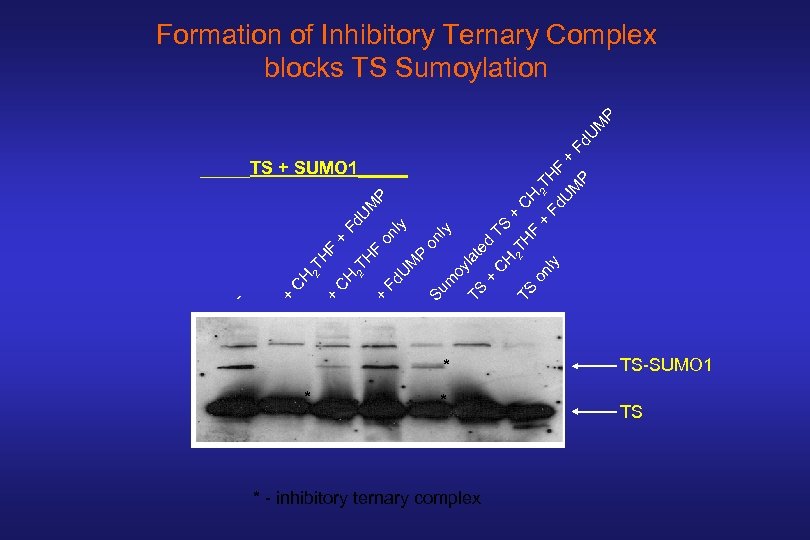 HF + * * - inhibitory ternary complex TS on ly 2 T TS CH te d + la TS oy Su m M P on l y on ly + Fd U H F 2 T H C + + - CH 2 T HF + Fd UM P _____TS + SUMO 1_____ C H + 2 TH Fd UM F + P Fd. U M P Formation of Inhibitory Ternary Complex blocks TS Sumoylation TS-SUMO 1 TS
HF + * * - inhibitory ternary complex TS on ly 2 T TS CH te d + la TS oy Su m M P on l y on ly + Fd U H F 2 T H C + + - CH 2 T HF + Fd UM P _____TS + SUMO 1_____ C H + 2 TH Fd UM F + P Fd. U M P Formation of Inhibitory Ternary Complex blocks TS Sumoylation TS-SUMO 1 TS
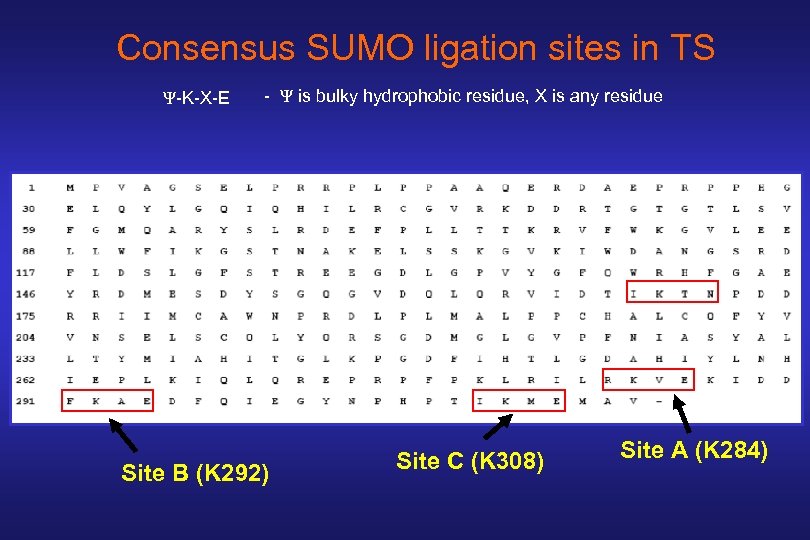 Consensus SUMO ligation sites in TS Ψ-K-X-E - Ψ is bulky hydrophobic residue, X is any residue Site B (K 292) Site C (K 308) Site A (K 284)
Consensus SUMO ligation sites in TS Ψ-K-X-E - Ψ is bulky hydrophobic residue, X is any residue Site B (K 292) Site C (K 308) Site A (K 284)
 281 313 ● Human TS: …ILRKVEKIDDFKAEDFQIEGYNPHPTIKMEMAV Mouse TS: T V Rat TS: T V
281 313 ● Human TS: …ILRKVEKIDDFKAEDFQIEGYNPHPTIKMEMAV Mouse TS: T V Rat TS: T V
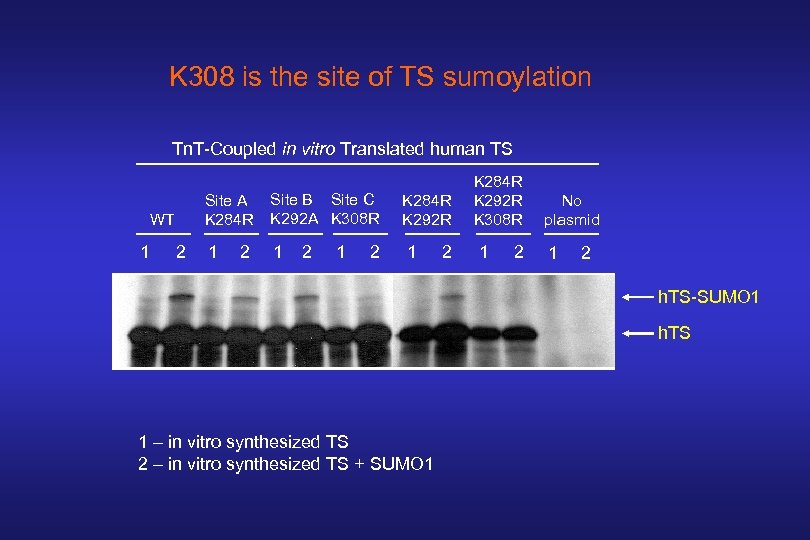 K 308 is the site of TS sumoylation Tn. T-Coupled in vitro Translated human TS Site A K 284 R WT 1 2 Site B Site C K 292 A K 308 R K 284 R K 292 R 1 1 1 2 2 K 284 R K 292 R K 308 R 1 2 No plasmid 1 2 h. TS-SUMO 1 h. TS 1 – in vitro synthesized TS 2 – in vitro synthesized TS + SUMO 1
K 308 is the site of TS sumoylation Tn. T-Coupled in vitro Translated human TS Site A K 284 R WT 1 2 Site B Site C K 292 A K 308 R K 284 R K 292 R 1 1 1 2 2 K 284 R K 292 R K 308 R 1 2 No plasmid 1 2 h. TS-SUMO 1 h. TS 1 – in vitro synthesized TS 2 – in vitro synthesized TS + SUMO 1
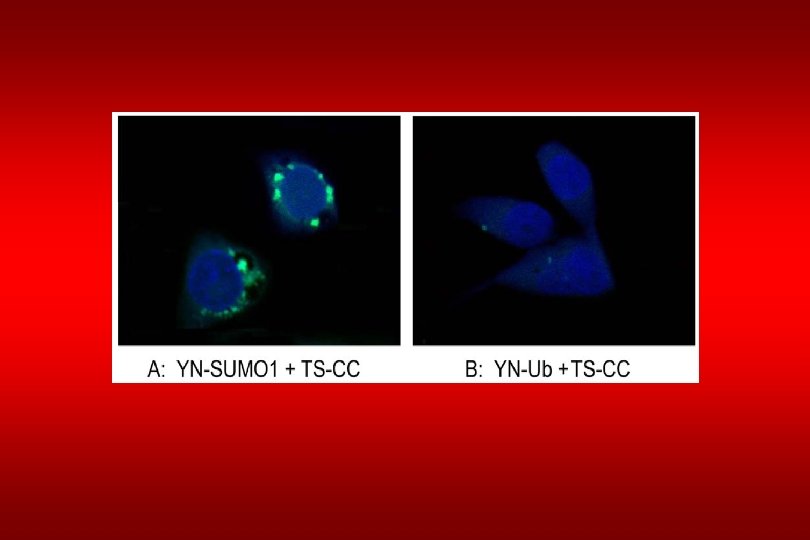
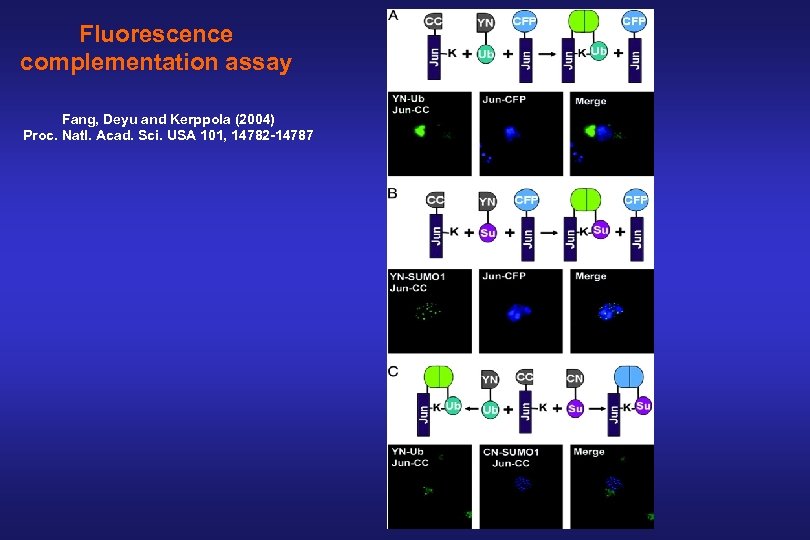 Fluorescence complementation assay Fang, Deyu and Kerppola (2004) Proc. Natl. Acad. Sci. USA 101, 14782 -14787
Fluorescence complementation assay Fang, Deyu and Kerppola (2004) Proc. Natl. Acad. Sci. USA 101, 14782 -14787
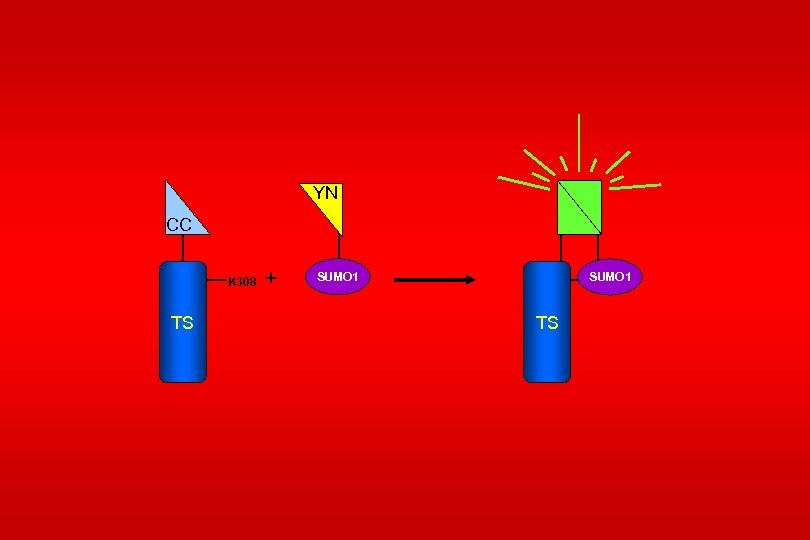 YN CC K 308 TS + SUMO 1 TS
YN CC K 308 TS + SUMO 1 TS
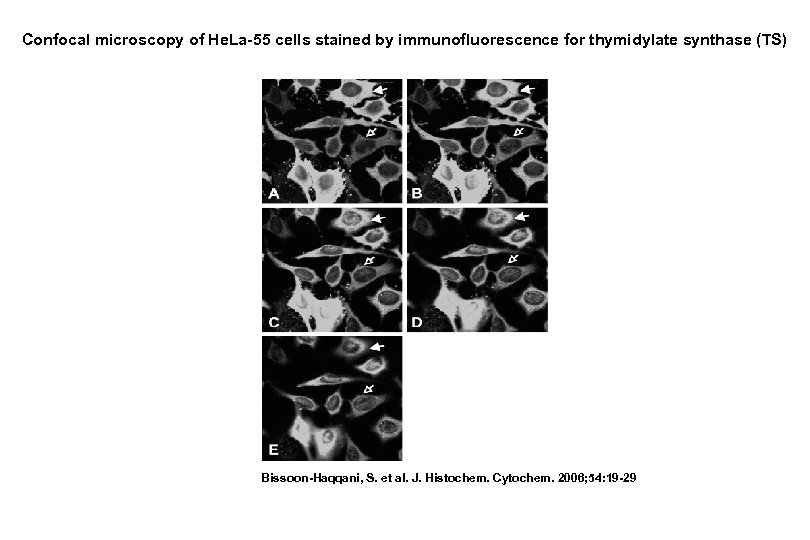 Confocal microscopy of He. La-55 cells stained by immunofluorescence for thymidylate synthase (TS) Bissoon-Haqqani, S. et al. J. Histochem. Cytochem. 2006; 54: 19 -29
Confocal microscopy of He. La-55 cells stained by immunofluorescence for thymidylate synthase (TS) Bissoon-Haqqani, S. et al. J. Histochem. Cytochem. 2006; 54: 19 -29
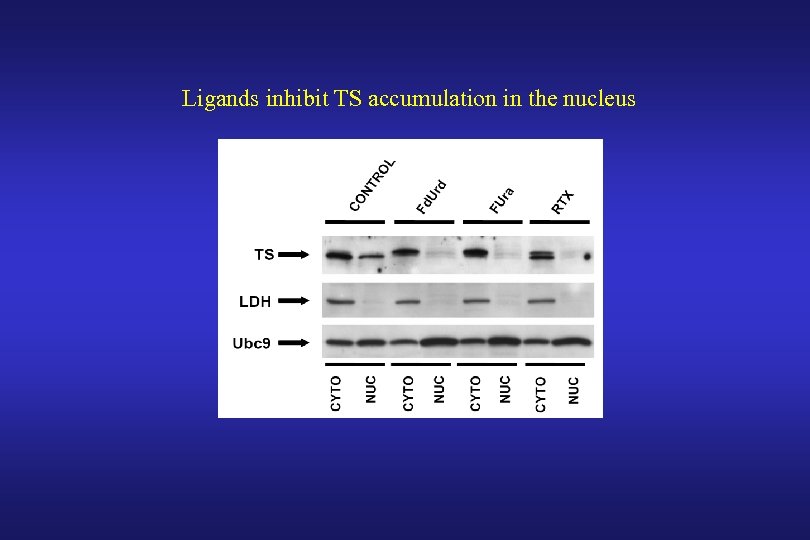 Ligands inhibit TS accumulation in the nucleus
Ligands inhibit TS accumulation in the nucleus
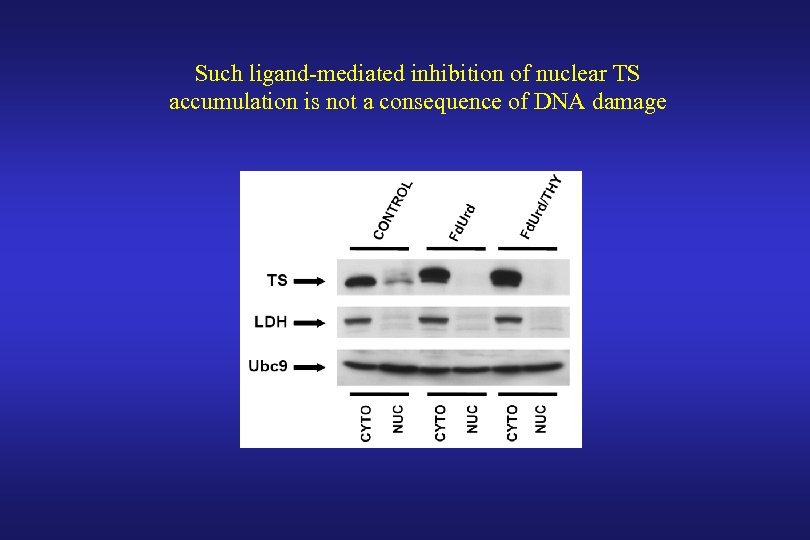 Such ligand-mediated inhibition of nuclear TS accumulation is not a consequence of DNA damage
Such ligand-mediated inhibition of nuclear TS accumulation is not a consequence of DNA damage
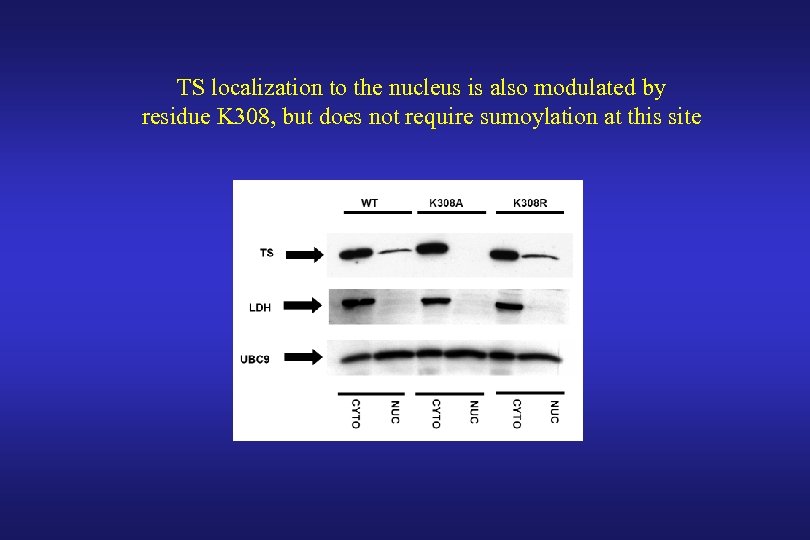 TS localization to the nucleus is also modulated by residue K 308, but does not require sumoylation at this site
TS localization to the nucleus is also modulated by residue K 308, but does not require sumoylation at this site
 “Working” Model for nuclear import of TS: (1. ) C-terminal end (including K 308) of TS is required for its binding to the nuclear periphery. (2. ) This is abrogated by either ligands or the K 308 A mutation. (3. ) TS goes through a sumoylation/desumoylation cycle as it enters the nucleus. (4. ) This determines the enzyme’s intra-nuclear locale. Ligands affect exposure of C-terminal region; Sumoylation affects intra-nuclear locale, but not entry.
“Working” Model for nuclear import of TS: (1. ) C-terminal end (including K 308) of TS is required for its binding to the nuclear periphery. (2. ) This is abrogated by either ligands or the K 308 A mutation. (3. ) TS goes through a sumoylation/desumoylation cycle as it enters the nucleus. (4. ) This determines the enzyme’s intra-nuclear locale. Ligands affect exposure of C-terminal region; Sumoylation affects intra-nuclear locale, but not entry.
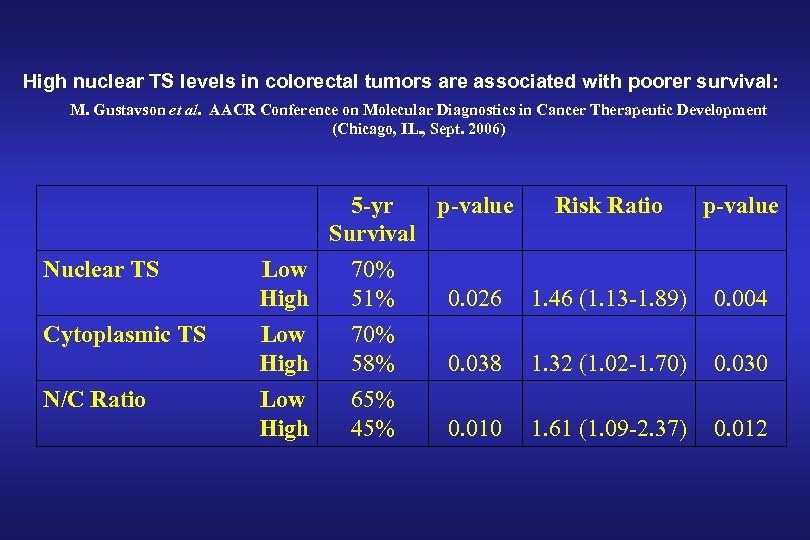 High nuclear TS levels in colorectal tumors are associated with poorer survival: M. Gustavson et al. AACR Conference on Molecular Diagnostics in Cancer Therapeutic Development (Chicago, IL. , Sept. 2006) 5 -yr p-value Survival Risk Ratio p-value Nuclear TS Low High 70% 51% 0. 026 1. 46 (1. 13 -1. 89) 0. 004 Cytoplasmic TS Low High 70% 58% 0. 038 1. 32 (1. 02 -1. 70) 0. 030 N/C Ratio Low High 65% 45% 0. 010 1. 61 (1. 09 -2. 37) 0. 012
High nuclear TS levels in colorectal tumors are associated with poorer survival: M. Gustavson et al. AACR Conference on Molecular Diagnostics in Cancer Therapeutic Development (Chicago, IL. , Sept. 2006) 5 -yr p-value Survival Risk Ratio p-value Nuclear TS Low High 70% 51% 0. 026 1. 46 (1. 13 -1. 89) 0. 004 Cytoplasmic TS Low High 70% 58% 0. 038 1. 32 (1. 02 -1. 70) 0. 030 N/C Ratio Low High 65% 45% 0. 010 1. 61 (1. 09 -2. 37) 0. 012
 Could the cytotoxicity of TS inhibitors derive from abrogation of the enzyme’s nuclear accumulation, and subsequent effects on DNA repair?
Could the cytotoxicity of TS inhibitors derive from abrogation of the enzyme’s nuclear accumulation, and subsequent effects on DNA repair?

 Karen Barbour Marj Peña Sandra Melo Kenn White Yang Xing
Karen Barbour Marj Peña Sandra Melo Kenn White Yang Xing


