8124ae2a7f6762e7ced70503e302ef74.ppt
- Количество слайдов: 31

Sys. Mo-DB: Towards “just enough” data exchange for the Sys. MO Consortium Carole Goble, Uni of Manchester, UK Jacky Snoep, Uni of Manchester, UK / Stellenbosch, South Africa Isabel Rojas, EML Research g. Gmb. H, Germany

l Pan European collaboration. l Systems Biology of Microorganisms. l The transition from growing to non-growing Bacillus subtilis cells Energy and Saccharomyces cerevisiae Biology of Clostridium acetobutylicum Gene interaction networks and models of cation homeostasis in Saccharomyces cerevisiae http: //www. sysmo. net l l l

l Eleven individual projects, 91 institutes l Different research outcomes l A cross-section of microorganisms, incl. bacteria, archaea and yeast. l Record and describe the dynamic molecular processes occurring in microorganisms in a comprehensive way l Present these processes in the form of computerized mathematical models. l Pool research capacities and know-how. l Already running since April 2007. Runs for 3 -5 years. l http: //www. sysmo. net Ba. Cell-Sys. MO COSMIC SUMO KOSMOBAC Sys. MO-LAB PSYSMO Valla MOSES TRANSLUCENT STREAM Sulfo. SYS
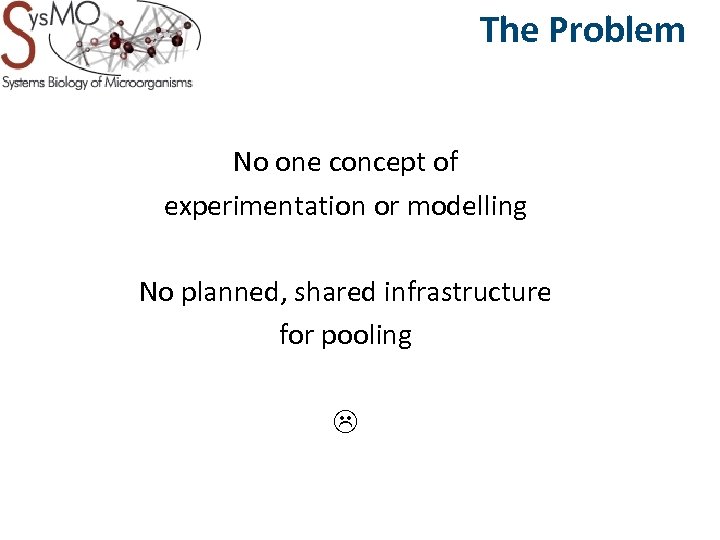
The Problem No one concept of experimentation or modelling No planned, shared infrastructure for pooling

Own solutions Own data solutions and collaboration environments. wikis, e-Groupware, PHProjekt, Base. Camp, PLONE, Alfresco, bespoke commercial … files and spreadsheets. Suspicion and caution over sharing. Interesting interplay between modellers, experimentalists and bioinformaticians. Data issues Many do not have data, or follow the standards that exist or know who is doing what. Much of the data cannot be compared Different organisms, different strains. Resource Issues No extra resources for the consortiums 91 institutes, 11 consortiums, some overlapping

DB Sys. MO-DB l Started July 2008, 3 years, 3+3 people, 3 teams over 3 sites l Sensitively retrofit a data access, model handling and data integration platform. l Support and manage the diversity of data, models and competencies. l Web-based solution: l exchange of data, models and processes (intra- and inter-consortia). l search for data, models and processes across the initiative. l dissemination of results.
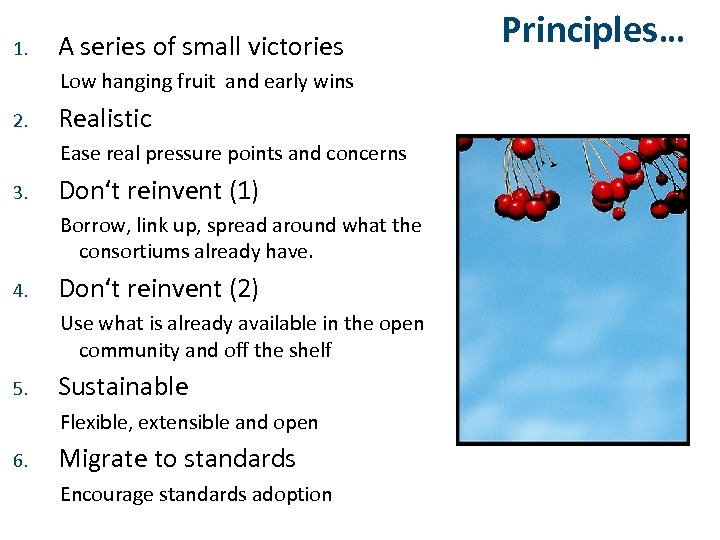
1. A series of small victories Low hanging fruit and early wins 2. Realistic Ease real pressure points and concerns 3. Don‘t reinvent (1) Borrow, link up, spread around what the consortiums already have. 4. Don‘t reinvent (2) Use what is already available in the open community and off the shelf 5. Sustainable Flexible, extensible and open 6. Migrate to standards Encourage standards adoption Principles…
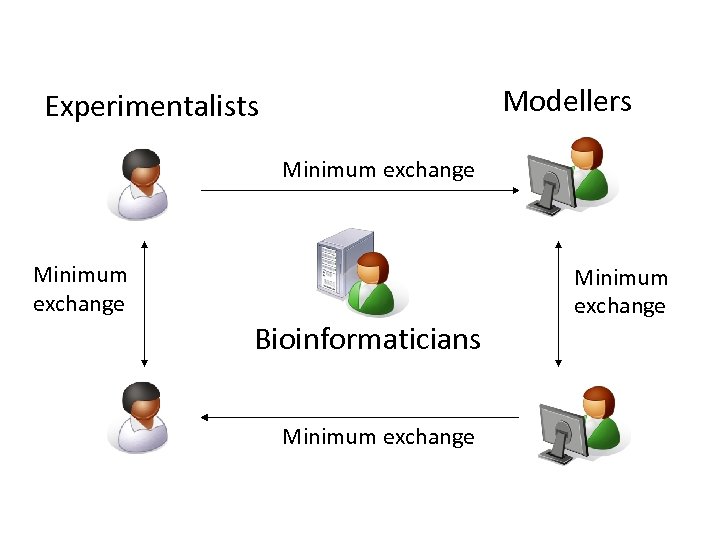
Modellers Experimentalists Minimum exchange Bioinformaticians Minimum exchange

Social Approach l Questionnaires l l PALS l l l Ranked projects Bronze, Silver, Gold and Platinum 18 Postdocs and Ph. D students All three kinds of people Our design and technical collaboration team Very intense face to face and virtual collaboration UK and Continental PALS Chapters Audits and Sharing l Methods, data, models, standards, software, schemas, spreadsheets, SOPs…. .
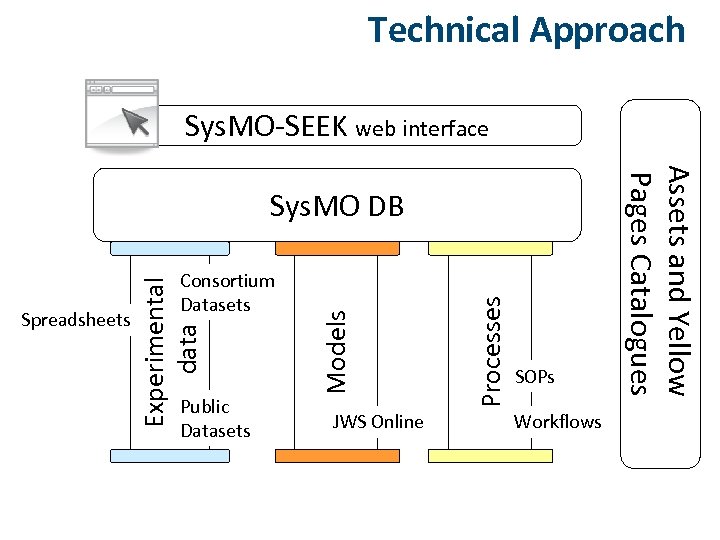
Technical Approach Sys. MO-SEEK web interface JWS Online Processes Public Datasets Models Experimental data Spreadsheets Consortium Datasets SOPs Workflows Assets and Yellow Pages Catalogues Sys. MO DB
Discovery Sys. MO-SEEK l Single, web based, access point l Single sign-on access control & versioning management l Single search point over yellow pages and assets catalogue l People, Expertise, SOP, Equipment l Metadata about Data – spreadsheets and databases l Models (JWS Online), workflows (my. Experiment), public web services (Bio. Catalogue) l Call out to external resources (e. g. Pub. Med) Does not hold results; holds metadata on results and links to results – pilot COSMIC consortium A component for Sys. MO groups to incorporate in their own environments and applications

Sys. MO SEEK (20 questions) Is there any group generating kinetic data? Is this data available? Who is working with which organism? ? ? What methods are been used to determine enzyme activity? Under which experimental conditions are my partners working on for the measurement of glucose concentration?
Models Publish, manage, run, validate SBML models l l Database of curated models and a model simulator Web service enabled to run from workflows Separate password protected websites for each project Through SEEK…. l l l Special instance of JWS Online for Sys. MO Validate and run models from Sys. MO-SEEK and publish later. Access control as do for other assets Access to other resources (Biomodels, Copasi) Semantic SBML from TRANSLUCENT project SBML and MIRIAM education
Experimental Processes l l l Protocols and SOPs assets deposited or linked to SOP gathering Nature Protocols format recommendation High level classification for indexing and tagging Got a few, need more.
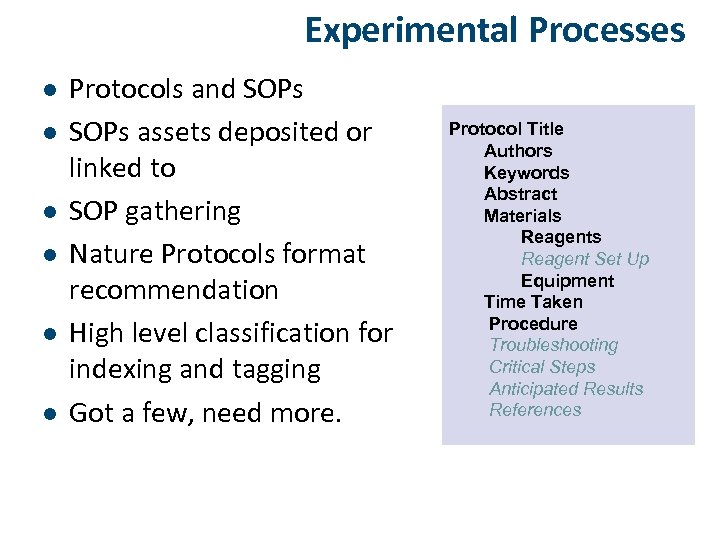
Experimental Processes l l l Protocols and SOPs assets deposited or linked to SOP gathering Nature Protocols format recommendation High level classification for indexing and tagging Got a few, need more. Protocol Title Authors Keywords Abstract Materials Reagent Set Up Equipment Time Taken Procedure Troubleshooting Critical Steps Anticipated Results References
Experimental Processes Deposition
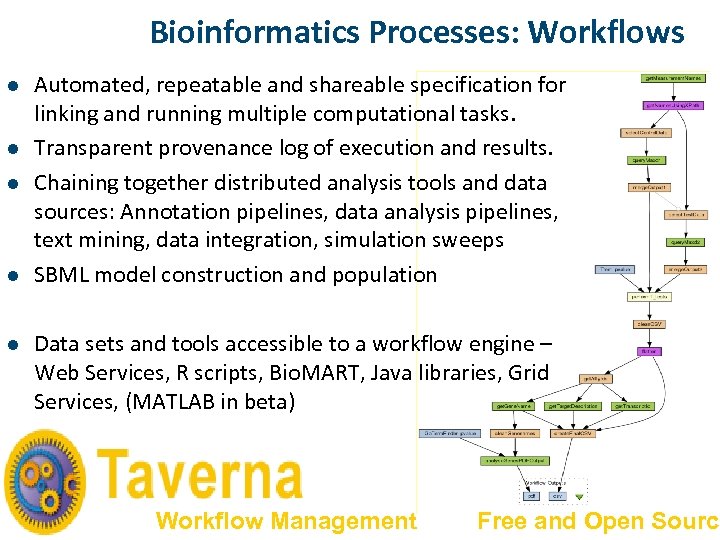
Bioinformatics Processes: Workflows l l l Automated, repeatable and shareable specification for linking and running multiple computational tasks. Transparent provenance log of execution and results. Chaining together distributed analysis tools and data sources: Annotation pipelines, data analysis pipelines, text mining, data integration, simulation sweeps SBML model construction and population Data sets and tools accessible to a workflow engine – Web Services, R scripts, Bio. MART, Java libraries, Grid Services, (MATLAB in beta) Workflow Management Free and Open Source
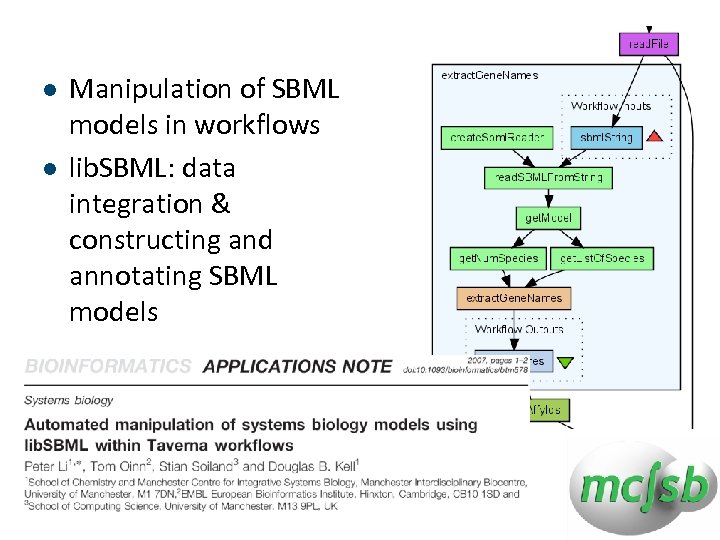
l l Manipulation of SBML models in workflows lib. SBML: data integration & constructing and annotating SBML models
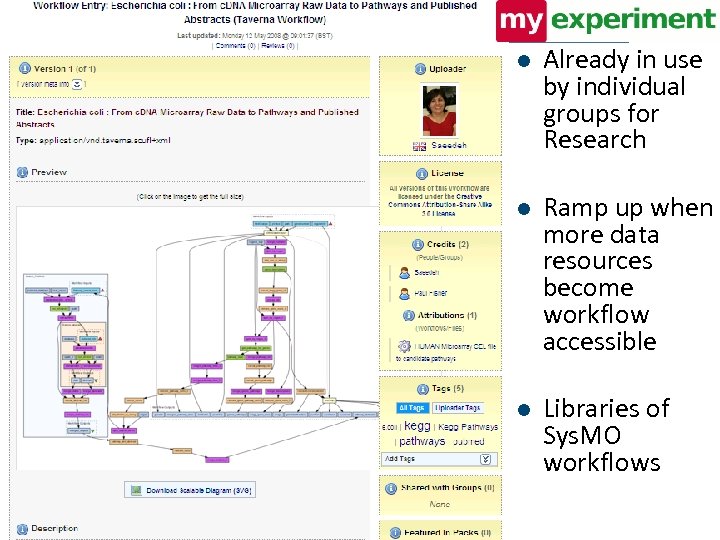
l Already in use by individual groups for Research l Ramp up when more data resources become workflow accessible l Libraries of Sys. MO workflows
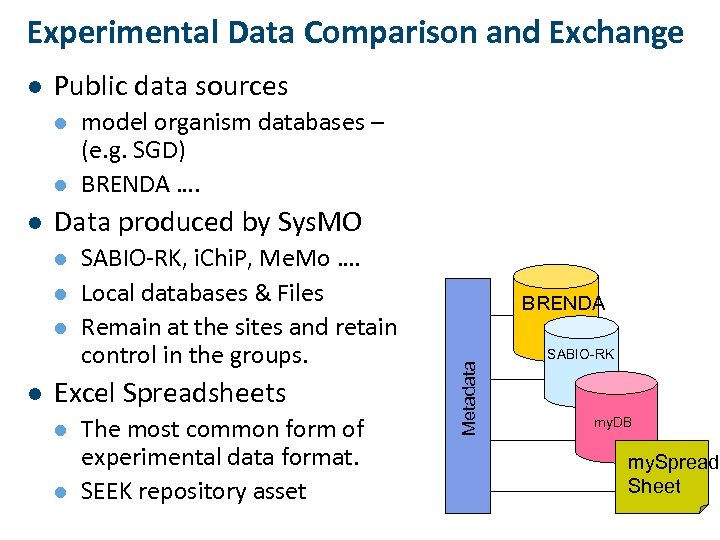
Experimental Data Comparison and Exchange Public data sources l l l Data produced by Sys. MO l l model organism databases – (e. g. SGD) BRENDA …. SABIO-RK, i. Chi. P, Me. Mo …. Local databases & Files Remain at the sites and retain control in the groups. Excel Spreadsheets l l The most common form of experimental data format. SEEK repository asset BRENDA Metadata l SABIO-RK my. DB my. Spread Sheet
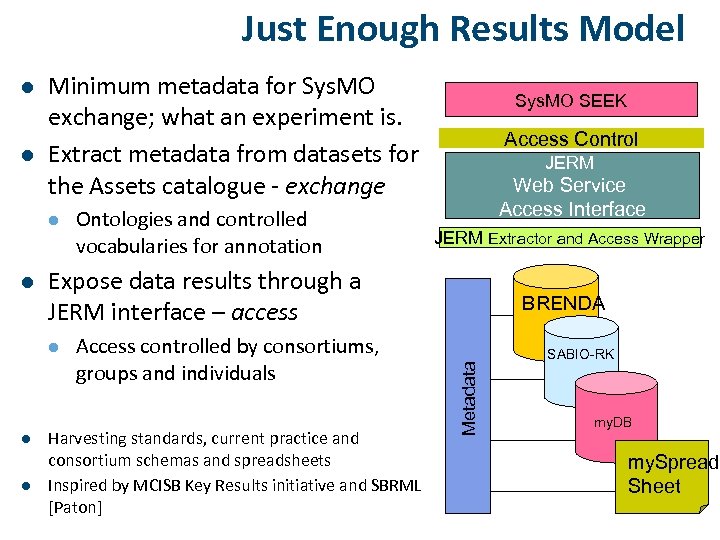
Just Enough Results Model l Minimum metadata for Sys. MO exchange; what an experiment is. Extract metadata from datasets for the Assets catalogue - exchange l l l Access Control JERM Web Service Access Interface JERM Extractor and Access Wrapper Expose data results through a JERM interface – access l l Ontologies and controlled vocabularies for annotation Sys. MO SEEK Access controlled by consortiums, groups and individuals Harvesting standards, current practice and consortium schemas and spreadsheets Inspired by MCISB Key Results initiative and SBRML [Paton] BRENDA Metadata l SABIO-RK my. DB my. Spread Sheet
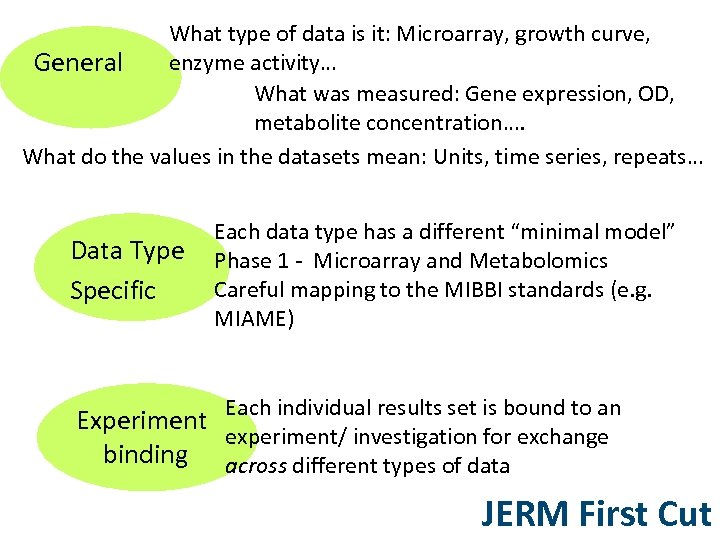
What type of data is it: Microarray, growth curve, enzyme activity… General What was measured: Gene expression, OD, metabolite concentration…. What do the values in the datasets mean: Units, time series, repeats… Data Type Specific Each data type has a different “minimal model” Phase 1 - Microarray and Metabolomics Careful mapping to the MIBBI standards (e. g. MIAME) Each individual results set is bound to an Experiment experiment/ investigation for exchange binding across different types of data JERM First Cut
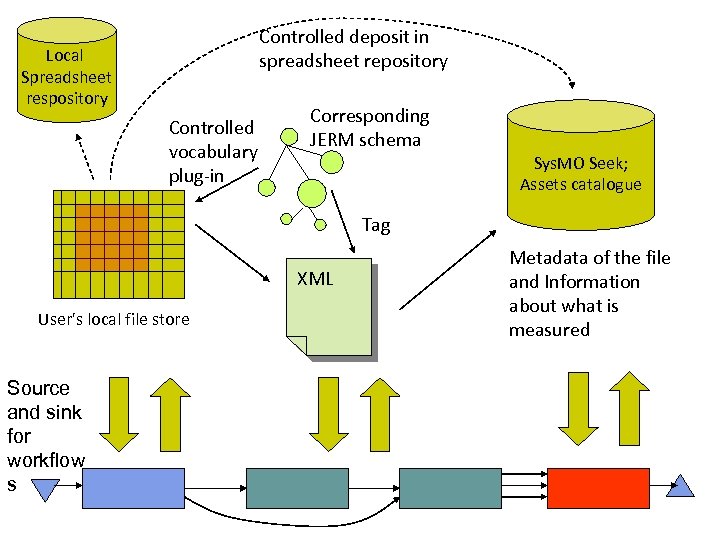
Controlled deposit in spreadsheet repository Local Spreadsheet respository Controlled vocabulary plug-in Corresponding JERM schema Sys. MO Seek; Assets catalogue Tag XML User's local file store Source and sink for workflow s Metadata of the file and Information about what is measured
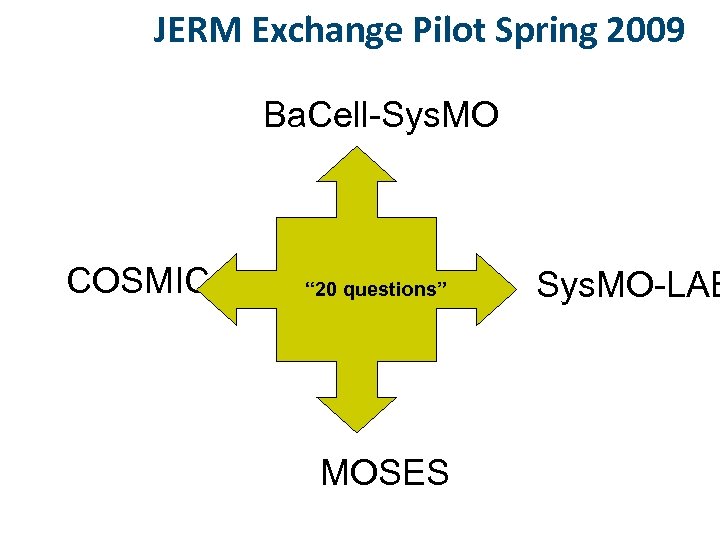
JERM Exchange Pilot Spring 2009 Ba. Cell-Sys. MO COSMIC “ 20 questions” MOSES Sys. MO-LAB
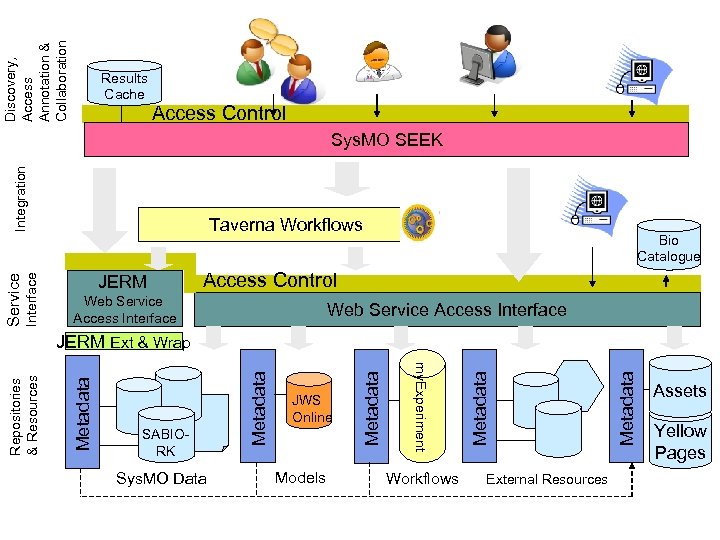
Discovery, Access Annotation & Collaboration Results Cache Access Control Interface Service Integration Sys. MO SEEK Taverna Workflows JERM Bio Catalogue Access Control Web Service Access Interface Sys. MO Data Models Workflows External Resources Metadata Metadata SABIORK JWS Online my. Experiment Repositories & Resources JERM Ext & Wrap Assets Yellow Pages
Related initiatives and sources l l l Open. Wet. Ware Cold Spring Harbor Protocols MIBBI National Centre for Bio. Ontologies OBO Foundary l Wikipathways Pathway commons Straininfo ONDEX l Pubmed l l l

Training and Know-how l Sys. MO-DB l l Training on databases, models, workflow systems and web services, and best practice for the annotation of resources by metadata. Kick-starting toolkits, workflows and SOP templates Summer schools Sys. MO consortium (esp. PALS) l l l Social networking for shared content, know-how and best practice Contribution Best of breed solutions in place already

Summary l Sys. MO-DB is an exercise in: l Sensitively retrofitting a data access, model handling and data integration platform. Supporting the diversity of data, models and competencies l l Social mediation and manipulation l Towards Just Enough™ exchange

Acknowledgements l l l Sys. MO-DB Team Sys. MO-PALS my. Grid, EML and JWS Online teams OMII-UK, Uni Southampton EBI, MCISB

Links l my. Experiment: http: //www. myexperiment. org Taverna: http: //www. mygrid. org. uk l JWS Online: http: //jjj. biochem. sun. ac. za/ l SABIO-RK http: //sabio. villa-bosch. de/ l
8124ae2a7f6762e7ced70503e302ef74.ppt