564a49a23236dc2a33898fd2009ef94e.ppt
- Количество слайдов: 42

Swift Fast, Reliable, Loosely Coupled Parallel Computation Ian Foster Computation Institute Argonne National Laboratory University of Chicago Joint work with Yong Zhao, Ioan Raicu, Mike Wilde, Ben Clifford, Mihael Hatigan, Tibi Stef-Praun, Veronika Nefedova
Case Study: The Functional MRI (f. MRI) Data Center l Online repository of neuroimaging data u u l A typical study comprises 3 groups, 20 subjects/group, 5 runs/subject, 300 volumes/run 90, 000 volumes, 60 GB raw 1. 2 million files processed 100 s of such studies in total Many users analyze this data u Wide range of analyses u Testing production u Ensembles: a set of data analyses by parameters, datasets 2
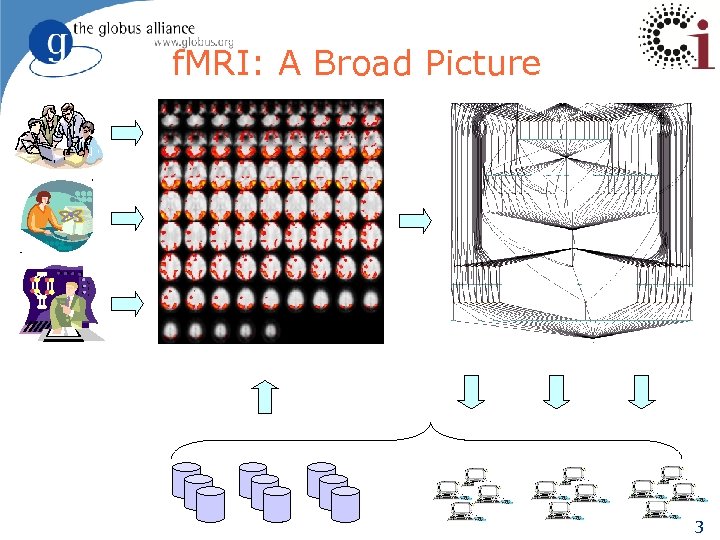
f. MRI: A Broad Picture 3

Challenges l Deluge of data: instrumentation, simulation l Data analysis turns into data integration l Community-wide collaboration l Provenance: tracking, query, application l Scalability: desktop to Grid l Productivity: throughput, performance 4
Swift System l Clean separation of logical/physical concerns u XDTM specification of logical data structures + Concise specification of parallel programs u Swift. Script, with iteration, etc. + Efficient execution on distributed resources u Karajan threading, Falkon provisioning, Globus interfaces, pipelining, load balancing + Rigorous provenance tracking and query u Virtual data schema & automated recording Improved usability and productivity u Demonstrated in numerous applications 5
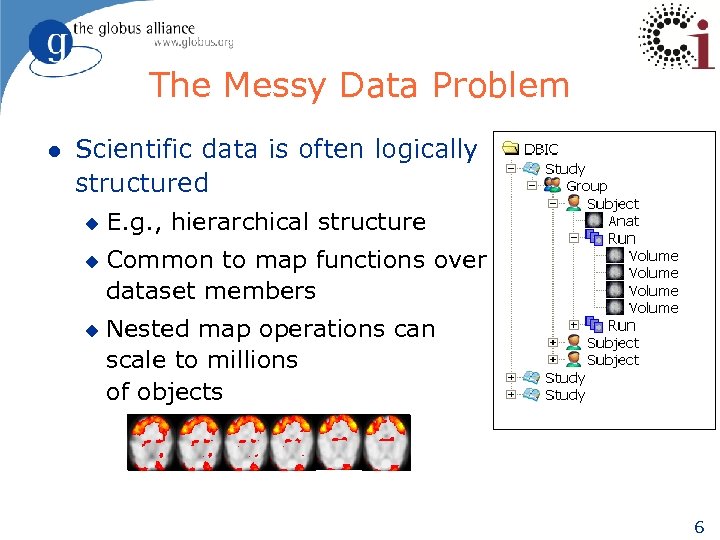
The Messy Data Problem l Scientific data is often logically structured u u u E. g. , hierarchical structure Common to map functions over dataset members Nested map operations can scale to millions of objects 6
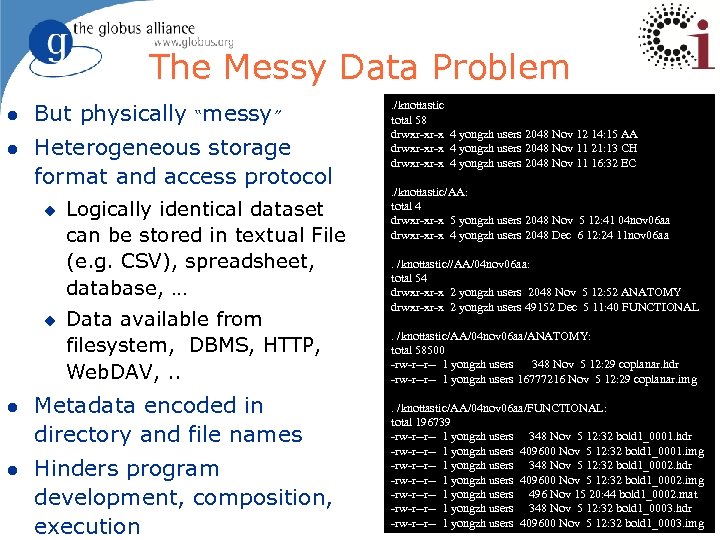
The Messy Data Problem l But physically “messy” l Heterogeneous storage format and access protocol u u Logically identical dataset can be stored in textual File (e. g. CSV), spreadsheet, database, … Data available from filesystem, DBMS, HTTP, Web. DAV, . . l Metadata encoded in directory and file names l Hinders program development, composition, execution . /knottastic total 58 drwxr-xr-x 4 yongzh users 2048 Nov 12 14: 15 AA drwxr-xr-x 4 yongzh users 2048 Nov 11 21: 13 CH drwxr-xr-x 4 yongzh users 2048 Nov 11 16: 32 EC. /knottastic/AA: total 4 drwxr-xr-x 5 yongzh users 2048 Nov 5 12: 41 04 nov 06 aa drwxr-xr-x 4 yongzh users 2048 Dec 6 12: 24 11 nov 06 aa. /knottastic//AA/04 nov 06 aa: total 54 drwxr-xr-x 2 yongzh users 2048 Nov 5 12: 52 ANATOMY drwxr-xr-x 2 yongzh users 49152 Dec 5 11: 40 FUNCTIONAL. /knottastic/AA/04 nov 06 aa/ANATOMY: total 58500 -rw-r--r-- 1 yongzh users 348 Nov 5 12: 29 coplanar. hdr -rw-r--r-- 1 yongzh users 16777216 Nov 5 12: 29 coplanar. img. /knottastic/AA/04 nov 06 aa/FUNCTIONAL: total 196739 -rw-r--r-- 1 yongzh users 348 Nov 5 12: 32 bold 1_0001. hdr -rw-r--r-- 1 yongzh users 409600 Nov 5 12: 32 bold 1_0001. img -rw-r--r-- 1 yongzh users 348 Nov 5 12: 32 bold 1_0002. hdr -rw-r--r-- 1 yongzh users 409600 Nov 5 12: 32 bold 1_0002. img -rw-r--r-- 1 yongzh users 496 Nov 15 20: 44 bold 1_0002. mat -rw-r--r-- 1 yongzh users 348 Nov 5 12: 32 bold 1_0003. hdr -rw-r--r-- 1 yongzh users 409600 Nov 5 12: 32 bold 1_0003. img 7
XML Dataset Typing & Mapping (XDTM) l Describe logical structure by XML Schema u u l Complex types (structs and arrays) Use mapping descriptors for mappings u u l Primitive scalar types: int, float, string, date, … How dataset elements are mapped to physical representations External parameters (e. g. location) Use XPath for dataset selection XDTM: XML Dataset Typing and Mapping for Specifying Datasets [EGC 05] 8
XDTM: Related Work l Data format standardization u l Data format description u u l FITS, CDF, HDF-5, DICOM DFDL [Beckerle, Westhead 04] embeds annotations with XML Schema PADS [Fisher, Gruber 05], PADX [Fernandez, Fisher 06], declarative specs of physical layout and semantic properties Logical object u u ADO [Microsoft 01], in memory relational model SDO [Beatty, Brodsky 03], logical data model for J 2 EE programming 9
XDTM: Implementation l Virtual integration u Each data source treated as virtual XML source u Data structure defined as XML schema u u l Mapper responsible for accessing source and translating to/from XML representation Bi-directional Common mapping interface u Data providers implement the interface l u Responsible for data access details Standard mapper implementations provided l String, file system, CSV, … 10
![Swift. Script l Typed parallel programming notation [SIGMOD 05, Springer 06] u u l Swift. Script l Typed parallel programming notation [SIGMOD 05, Springer 06] u u l](https://present5.com/presentation/564a49a23236dc2a33898fd2009ef94e/image-11.jpg)
Swift. Script l Typed parallel programming notation [SIGMOD 05, Springer 06] u u l XDTM as data model and type system Typed dataset and procedure definitions Scripting language u Implicit data parallelism Program composition Clean application logic Type checking from procedures Dataset selection, iteration u Control constructs Discovery by types (foreach, if, while, …) Type conversion u A Notation & System for Expressing and Executing Cleanly Typed Workflows on Messy Scientific Data [SIGMOD 05] 11
![Swift. Script: Related Work l Coordination language Linda[Ahuja, Carriero 86], Strand[Foster, Taylor 90], PCN[Foster Swift. Script: Related Work l Coordination language Linda[Ahuja, Carriero 86], Strand[Foster, Taylor 90], PCN[Foster](https://present5.com/presentation/564a49a23236dc2a33898fd2009ef94e/image-12.jpg)
Swift. Script: Related Work l Coordination language Linda[Ahuja, Carriero 86], Strand[Foster, Taylor 90], PCN[Foster 92] u Durra[Barbacci, Wing 86], MANIFOLD[Papadopoulos 98] u Components programmed in specific language (C, FORTRAN) and linked with system u l “Workflow” languages and systems Taverna[Oinn, Addis 04], Kepler[Ludäscher, Altintas 05], Triana [Churches, Gombas 05], Vistrail[Callahan, Freire 06], DAGMan, Star-P u XPDL[Wf. MC 02], BPEL[Andrews, Curbera 03], and BPML[BPML 02], YAWL[van de Aalst, Hofstede 05], Windows Workflow Foundation [Microsoft 05] u 12
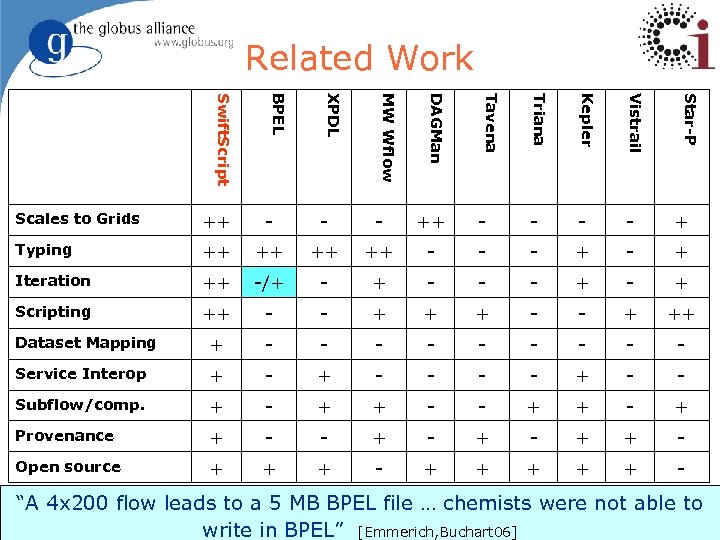
Related Work Kepler Vistrail Star-P Triana Tavena DAGMan MW Wflow XPDL BPEL Swift. Script Scales to Grids ++ - - - - + Typing ++ ++ - - - + Iteration ++ -/+ - - - + Scripting ++ - - + ++ Dataset Mapping + - - - - - Service Interop + - - - - + - - Subflow/comp. + - + Provenance + - - + - + + - Open source + + + - “A 4 x 200 flow leads to a 5 MB BPEL file … chemists were not able to 13 write in BPEL” [Emmerich, Buchart 06]
![f. MRI Type Definitions in Swift. Script type Study { Group g[ ]; } f. MRI Type Definitions in Swift. Script type Study { Group g[ ]; }](https://present5.com/presentation/564a49a23236dc2a33898fd2009ef94e/image-14.jpg)
f. MRI Type Definitions in Swift. Script type Study { Group g[ ]; } type Image {}; type Group { Subject s[ ]; } type Warp {}; type Subject { Volume anat; Run run[ ]; } type Run { Volume v[ ]; } Simplified version of f. MRI AIRSN Program (Spatial Normalization) type Volume { Image img; Header hdr; } type Header {}; type Air {}; type Air. Vec { Air a[ ]; } type Norm. Anat { Volume anat; Warp a. Warp; Volume n. Hires; } 14
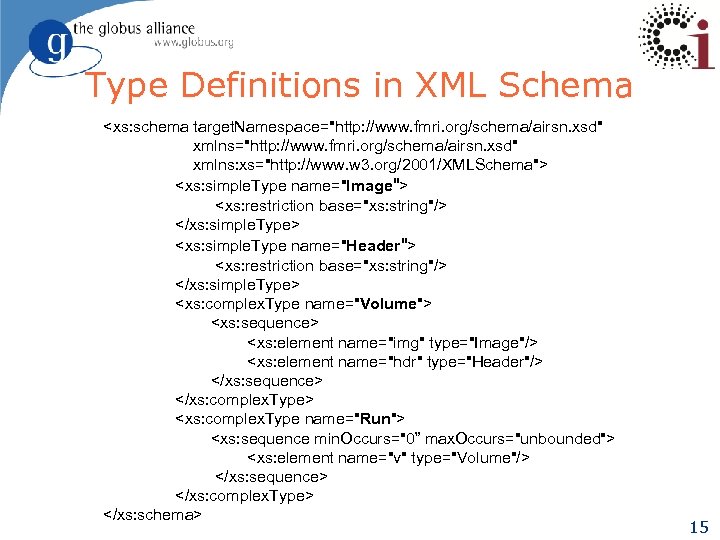
Type Definitions in XML Schema <xs: schema target. Namespace="http: //www. fmri. org/schema/airsn. xsd" xmlns: xs="http: //www. w 3. org/2001/XMLSchema"> <xs: simple. Type name="Image"> <xs: restriction base="xs: string"/> </xs: simple. Type> <xs: simple. Type name="Header"> <xs: restriction base="xs: string"/> </xs: simple. Type> <xs: complex. Type name="Volume"> <xs: sequence> <xs: element name="img" type="Image"/> <xs: element name="hdr" type="Header"/> </xs: sequence> </xs: complex. Type> <xs: complex. Type name="Run"> <xs: sequence min. Occurs="0” max. Occurs="unbounded"> <xs: element name="v" type="Volume"/> </xs: sequence> </xs: complex. Type> </xs: schema> 15

AIRSN Program Definition (Run or) reorient. Run (Run ir, string direction) { (Run snr) functional ( Run r, Norm. Anat a, foreach Volume iv, i in ir. v { Air shrink ) { or. v[i] = reorient(iv, direction); Run yro. Run = reorient. Run( r , "y" ); } Run ro. Run = reorient. Run( yro. Run , "x" ); } Volume std = ro. Run[0]; Run rndr = random_select( ro. Run, 0. 1 ); Air. Vector rnd. Air. Vec = align_linear. Run( rndr, std, 12, 1000, "81 3 3" ); Run resliced. Rndr = reslice. Run( rndr, rnd. Air. Vec, "o", "k" ); Volume mean. Rand = softmean( resliced. Rndr, "y", "null" ); Air mn. QAAir = alignlinear( a. n. Hires, mean. Rand, 6, 1000, 4, "81 3 3" ); Warp bold. Norm. Warp = combinewarp( shrink, a. a. Warp, mn. QAAir ); Run nr = reslice_warp_run( bold. Norm. Warp, ro. Run ); Volume mean. All = strictmean( nr, "y", "null" ) Volume bold. Mask = binarize( mean. All, "y" ); snr = gsmooth. Run( nr, bold. Mask, "6 6 6" ); 16
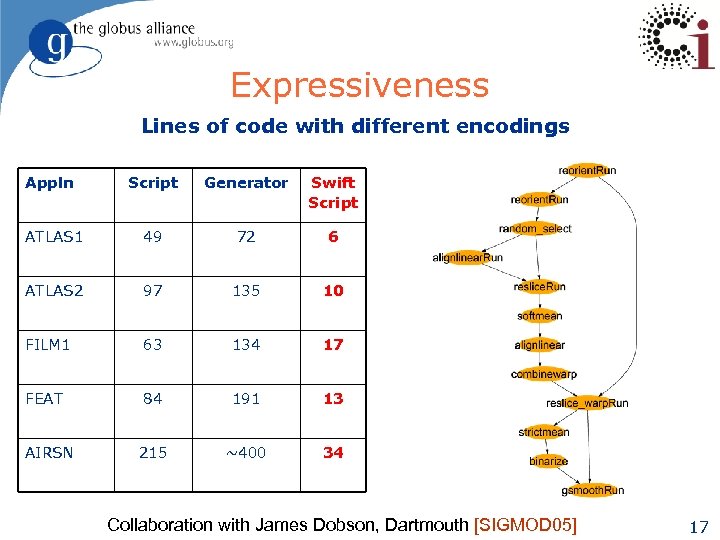
Expressiveness Lines of code with different encodings Appln Script Generator Swift Script ATLAS 1 49 72 6 ATLAS 2 97 135 10 FILM 1 63 134 17 FEAT 84 191 13 215 ~400 34 AIRSN Collaboration with James Dobson, Dartmouth [SIGMOD 05] 17
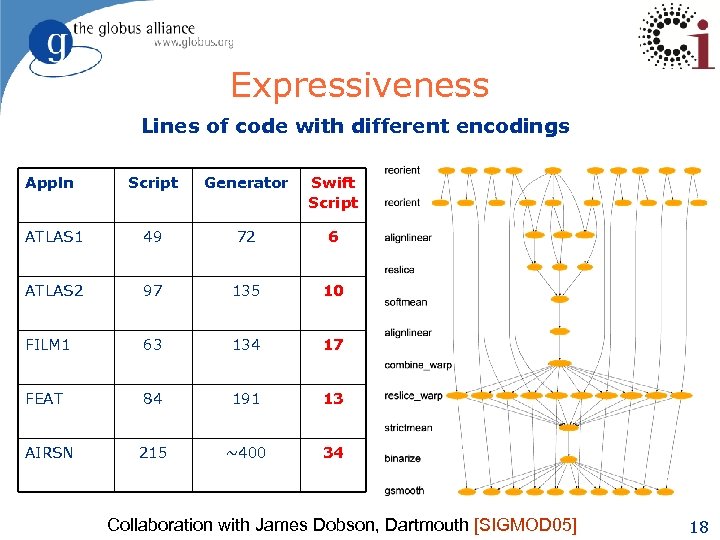
Expressiveness Lines of code with different encodings Appln Script Generator Swift Script ATLAS 1 49 72 6 ATLAS 2 97 135 10 FILM 1 63 134 17 FEAT 84 191 13 215 ~400 34 AIRSN Collaboration with James Dobson, Dartmouth [SIGMOD 05] 18

Dynamic Provisioning: Swift Architecture Specification Abstract computation Swift. Script Compiler Virtual Data Catalog Swift. Script Scheduling Execution Engine (Karajan w/ Swift Runtime) Virtual Node(s) C C Swift runtime callouts Status reporting Provenance collector Provisioning file 1 Falkon Resource Provisioner launcher App F 1 Provenance data launcher Provenance data file 2 App F 2 file 3 Amazon EC 2 Yong Zhao, Mihael Hatigan, Ioan Raicu, Mike Wilde, Ben Clifford
Swift Runtime System l Runtime system for Swift. Script [SSDBM 02, CIDR 03, Springer 06] u u Populate, update, query virtual data products Schedule, monitor, execute resulting computation on distributed Grid resources u u l Annotate virtual data products with customized metadata Trace provenance of virtual data products Grid scheduling and optimization u Lightweight execution engine: Karajan u Dynamic resource provisioning u Site selection, data movement, caching u Pipelining, clustering, load balancing u Fault tolerance, exception handling A Virtual Data System for Representing, Querying & Automating Data Derivation [SSDBM 02] 20
Swift uses Karajan Workflow Engine l Fast, scalable threading model l Suitable constructs for control flow l Flexible task dependency model u l “Futures” enable pipelining Flexible provider model allows for use of different run time environments u u l Job execution and data transfer Flow controlled to avoid resource overload Workflow client runs from a Java container Java Co. G Workflow, Gregor von Laszewski et al. , Workflows for Science, 2007 21
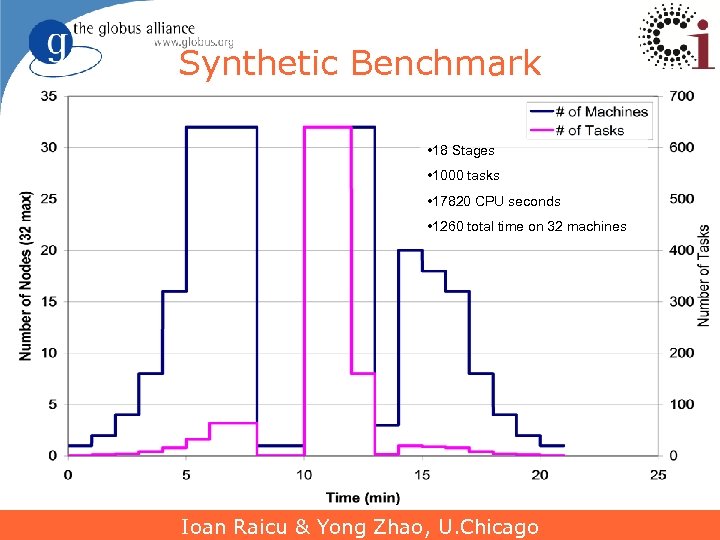
Synthetic Benchmark • 18 Stages • 1000 tasks • 17820 CPU seconds • 1260 total time on 32 machines Ioan Raicu & Yong Zhao, U. Chicago 22
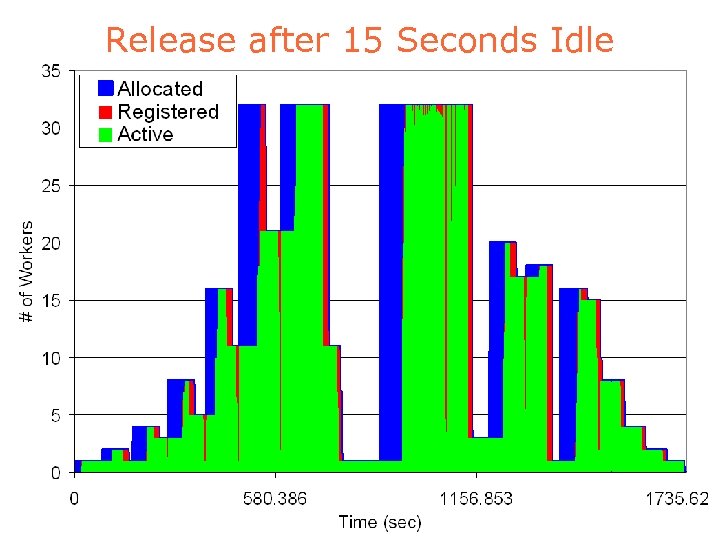
Release after 15 Seconds Idle
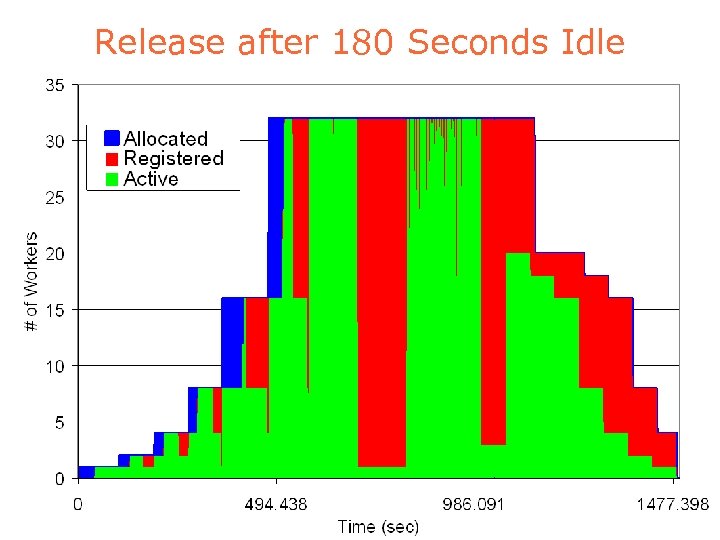
Release after 180 Seconds Idle

Swift Application B. Berriman, J. Good (Caltech) J. Jacob, D. Katz (JPL) 25
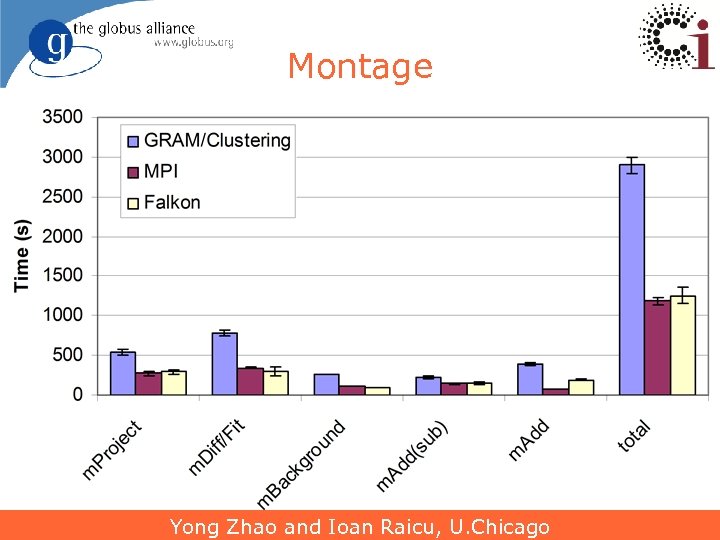
Montage Yong Zhao and Ioan Raicu, U. Chicago 26
Molecular Dynamics 27
28
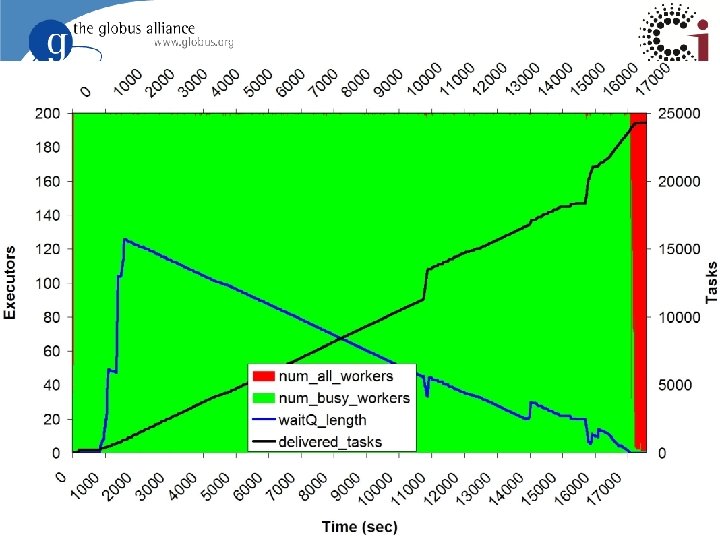
29

30

31
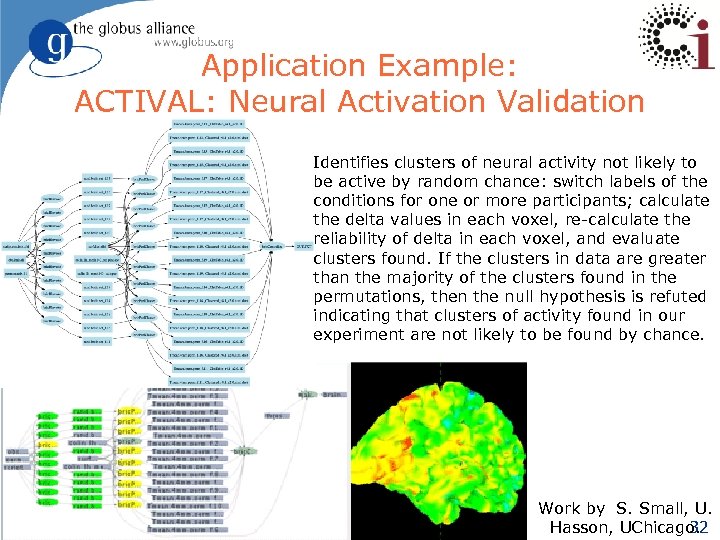
Application Example: ACTIVAL: Neural Activation Validation Identifies clusters of neural activity not likely to be active by random chance: switch labels of the conditions for one or more participants; calculate the delta values in each voxel, re-calculate the reliability of delta in each voxel, and evaluate clusters found. If the clusters in data are greater than the majority of the clusters found in the permutations, then the null hypothesis is refuted indicating that clusters of activity found in our experiment are not likely to be found by chance. Work by S. Small, U. Hasson, UChicago. 32

Swift. Script Program ACTIVAL – Datatypes & Utilities type type script {} type full. Brain. Data {} brain. Measurements{} type full. Brain. Specs {} precomputed. Permutations{} type brain. Dataset {} brain. Cluster. Table {} brain. Datasets{ brain. Dataset b[]; } brain. Clusters{ brain. Cluster. Table c[]; } // Procedure to run "R" statistical package (brain. Dataset t) bric. RInvoke (script permutation. Script, int iteration. No, brain. Measurements data. All, precomputed. Permutations data. Perm) { app { bric. RInvoke @filename(permutation. Script) iteration. No @filename(data. All) @filename(data. Perm); } } // Procedure to run AFNI Clustering tool (brain. Cluster. Table v, brain. Dataset t) bric. Cluster (script cluster. Script, int iteration. No, brain. Dataset rand. Brain, full. Brain. Data brain. File, full. Brain. Specs spec. File) { app { bric. Perl. Cluster @filename(cluster. Script) iteration. No @filename(rand. Brain) @filename(brain. File) @filename(spec. File); } } // Procedure to merge results based on statistical likelhoods (brain. Cluster. Table t) bric. Centralize ( brain. Cluster. Table bc[]) { app { bric. Centralize @filenames(bc); } } 33
ACTIVAL: Dataset Iteration Procedures // Procedure to iterate over the data collection (brain. Clusters rand. Cluster, brain. Datasets dset. Return) brain_cluster (full. Brain. Data brain. File, full. Brain. Specs spec. File) { int sequence[]=[1: 2000]; brain. Measurements precomputed. Permutations script brain. Datasets } data. All<fixed_mapper; file="obs. imit. all">; data. Perm<fixed_mapper; file="perm. matrix. 11">; rand. Script<fixed_mapper; file="script. obs. imit. tibi">; cluster. Script<fixed_mapper; file="surfclust. tibi">; rand. Brains<simple_mapper; prefix="rand. brain. set">; foreach int i in sequence { rand. Brains. b[i] = bric. RInvoke(rand. Script, i, data. All, data. Perm); brain. Dataset r. Brain=rand. Brains. b[i]; (rand. Cluster. c[i], dset. Return. b[i]) = bric. Cluster(cluster. Script, i, r. Brain, brain. File, spec. File); } 34

ACTIVAL: Main Program // Declare datasets full. Brain. Data full. Brain. Specs brain. File<fixed_mapper; file="colin_lh_mesh 140_std. pial. asc">; spec. File<fixed_mapper; file="colin_lh_mesh 140_std. spec">; brain. Datasets brain. Clusters rand. Brain<simple_mapper; prefix="rand. brain. set">; rand. Cluster<simple_mapper; prefix="Tmean. 4 mm. perm", suffix="_Clst. Table_r 4. 1_a 2. 0. 1 D">; dset. Return<simple_mapper; prefix="Tmean. 4 mm. perm", suffix="_Clustered_r 4. 1_a 2. 0. niml. dset">; cluster. Thresholds. Table<fixed_mapper; file="thresholds. table">; brain. Result<fixed_mapper; file="brain. final. dset">; orig. Brain<fixed_mapper; file="brain. permutation. 1">; brain. Datasets brain. Cluster. Table brain. Dataset // Main program – executes the entire application (rand. Cluster, dset. Return) = brain_cluster(brain. File, spec. File); cluster. Thresholds. Table = bric. Centralize (rand. Cluster. c); brain. Result = makebrain(orig. Brain, cluster. Thresholds. Table, brain. File, spec. File); 35
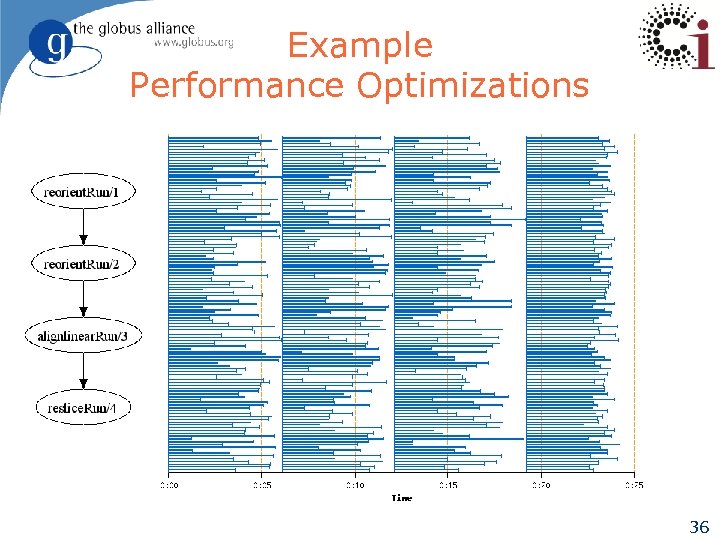
Example Performance Optimizations 36
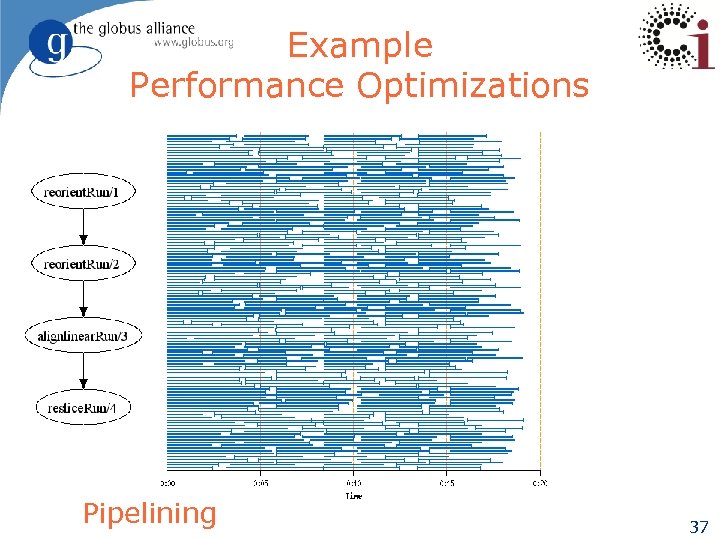
Example Performance Optimizations Pipelining 37
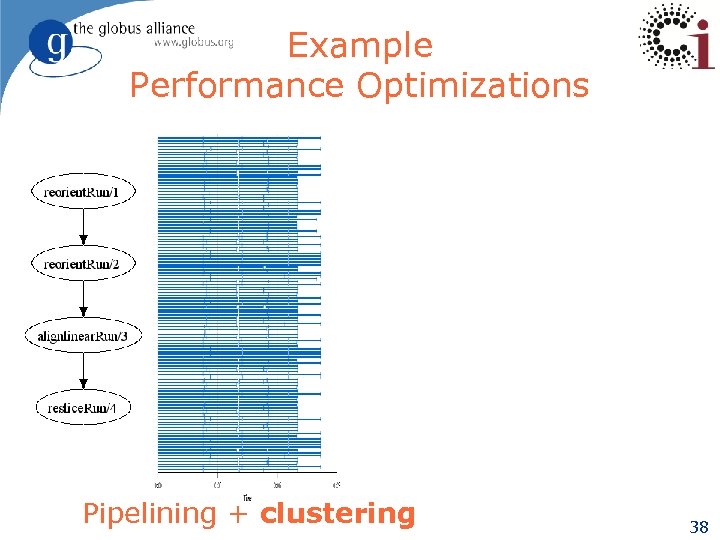
Example Performance Optimizations Pipelining + clustering 38
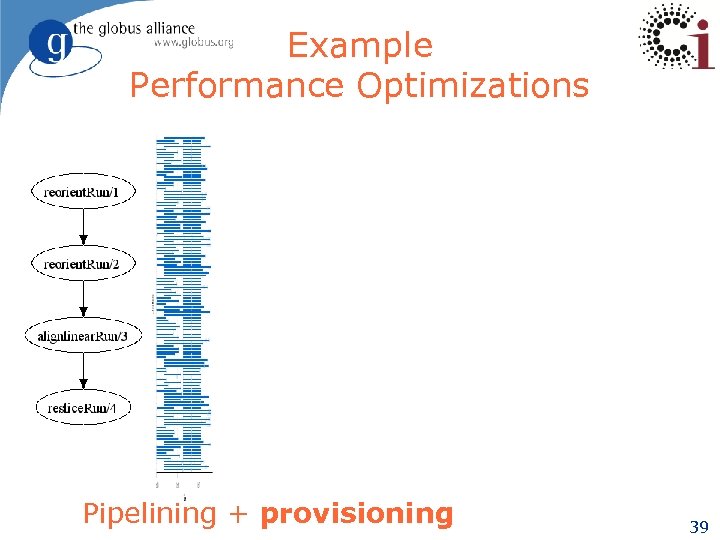
Example Performance Optimizations Pipelining + provisioning 39
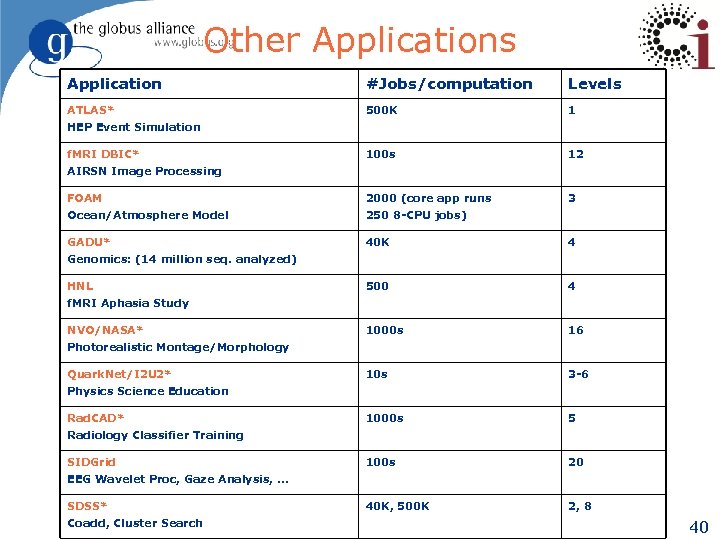
Other Applications Application #Jobs/computation Levels ATLAS* 500 K 1 100 s 12 FOAM 2000 (core app runs 3 Ocean/Atmosphere Model 250 8 -CPU jobs) GADU* 40 K 4 500 4 1000 s 16 10 s 3 -6 1000 s 5 100 s 20 40 K, 500 K 2, 8 HEP Event Simulation f. MRI DBIC* AIRSN Image Processing Genomics: (14 million seq. analyzed) HNL f. MRI Aphasia Study NVO/NASA* Photorealistic Montage/Morphology Quark. Net/I 2 U 2* Physics Science Education Rad. CAD* Radiology Classifier Training SIDGrid EEG Wavelet Proc, Gaze Analysis, … SDSS* Coadd, Cluster Search 40
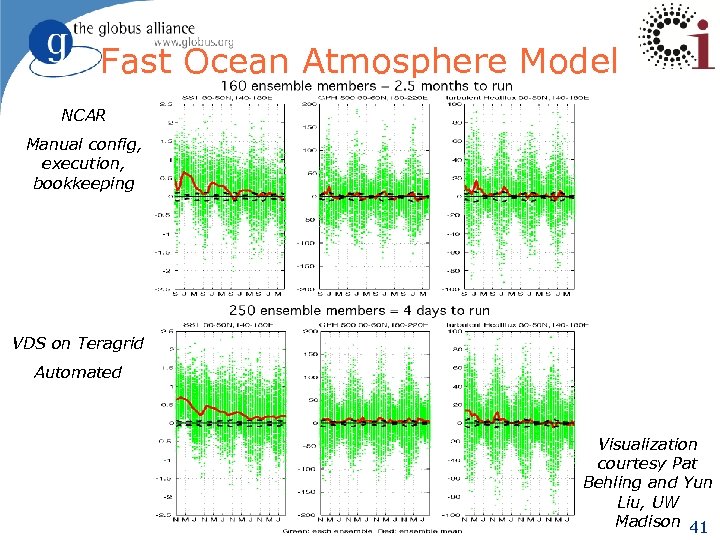
Fast Ocean Atmosphere Model NCAR Manual config, execution, bookkeeping VDS on Teragrid Automated Visualization courtesy Pat Behling and Yun Liu, UW Madison 41
Swift System www. ci. uchicago. edu/swift l Clean separation of logical/physical concerns u XDTM specification of logical data structures + Concise specification of parallel programs u Swift. Script, with iteration, etc. + Efficient execution on distributed resources u Lightweight threading, dynamic provisioning, Grid interfaces, pipelining, load balancing + Rigorous provenance tracking and query u Virtual data schema & automated recording Improved usability and productivity u Demonstrated in numerous applications 42
564a49a23236dc2a33898fd2009ef94e.ppt