362f40c8d9ad6f124fd8194d5d3cb731.ppt
- Количество слайдов: 21
 Supporting the Computation Needs of Structural Genomics Zach Miller Computer Sciences Department University of Wisconsin-Madison zmiller@cs. wisc. edu http: //www. cs. wisc. edu/condor
Supporting the Computation Needs of Structural Genomics Zach Miller Computer Sciences Department University of Wisconsin-Madison zmiller@cs. wisc. edu http: //www. cs. wisc. edu/condor
 Overview › What is structural genomics? › Problems we are trying to solve › Applications we use and how they › › interface with Condor Future work Conclusion www. cs. wisc. edu/condor
Overview › What is structural genomics? › Problems we are trying to solve › Applications we use and how they › › interface with Condor Future work Conclusion www. cs. wisc. edu/condor
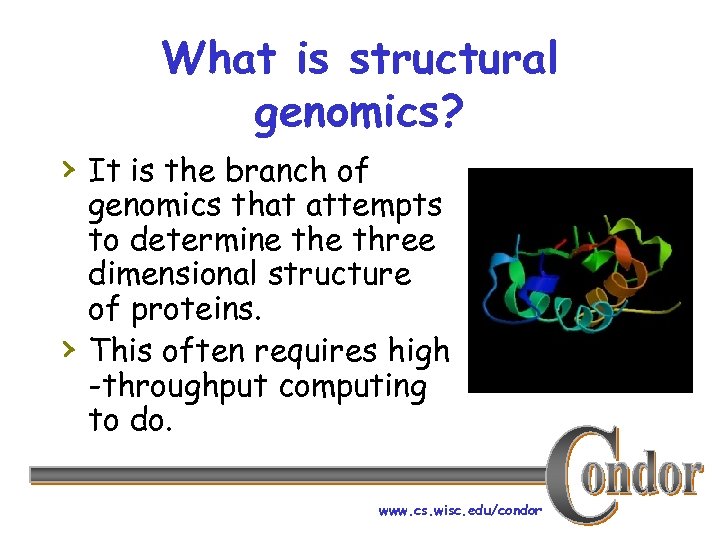 What is structural genomics? › It is the branch of › genomics that attempts to determine three dimensional structure of proteins. This often requires high -throughput computing to do. www. cs. wisc. edu/condor
What is structural genomics? › It is the branch of › genomics that attempts to determine three dimensional structure of proteins. This often requires high -throughput computing to do. www. cs. wisc. edu/condor
 Problems we are trying to solve › Target selection – which protein sequences are interesting and worth spending time calculating structures of? h. BLAST › Protein structure determination – what is the 3 D shape of a given protein sequence? h. CNS h. CYANA www. cs. wisc. edu/condor
Problems we are trying to solve › Target selection – which protein sequences are interesting and worth spending time calculating structures of? h. BLAST › Protein structure determination – what is the 3 D shape of a given protein sequence? h. CNS h. CYANA www. cs. wisc. edu/condor
 BLAST › BLAST is developed and supported by › › NCBI, part of the NIH. The NCBI BLAST home page is http: //www. ncbi. nlm. nih. gov/ BLAST is a search tool with special allowances for incomplete data and partial matches. www. cs. wisc. edu/condor
BLAST › BLAST is developed and supported by › › NCBI, part of the NIH. The NCBI BLAST home page is http: //www. ncbi. nlm. nih. gov/ BLAST is a search tool with special allowances for incomplete data and partial matches. www. cs. wisc. edu/condor
 BLAST target selection › By comparing different sets of whole › or partial sequences against other databases of known sequences, you can determine if the sequence you are trying to discover is already part of another database. In this way you can determine the interesting sequences to work on. www. cs. wisc. edu/condor
BLAST target selection › By comparing different sets of whole › or partial sequences against other databases of known sequences, you can determine if the sequence you are trying to discover is already part of another database. In this way you can determine the interesting sequences to work on. www. cs. wisc. edu/condor
 BLAST and Condor › Large BLAST searches are easily split › into smaller chunks that can be executed in parallel. There are two basic approaches: h. Split the input query into smaller chunks (our approach) h. Split the database into smaller chunks (mpi. BLAST approach) www. cs. wisc. edu/condor
BLAST and Condor › Large BLAST searches are easily split › into smaller chunks that can be executed in parallel. There are two basic approaches: h. Split the input query into smaller chunks (our approach) h. Split the database into smaller chunks (mpi. BLAST approach) www. cs. wisc. edu/condor
 BLAST and Condor › Doing thousands of queries against › multiple databases is easy using the Condor/BLAST framework. Features of the framework: h. Input queries can come from a file, ftp, or http h. Input queries can be in FASTA or XML format www. cs. wisc. edu/condor
BLAST and Condor › Doing thousands of queries against › multiple databases is easy using the Condor/BLAST framework. Features of the framework: h. Input queries can come from a file, ftp, or http h. Input queries can be in FASTA or XML format www. cs. wisc. edu/condor
 BLAST and Condor › More features of the framework: h. Databases can also be local files or automatically fetched via ftp or http and also in either FASTA or XML format h. Database Indexes can be automatically built using formatdb h. Multiple input files are joined or split as appropriate to fine-tune throughput h. Output can be delivered via ftp www. cs. wisc. edu/condor
BLAST and Condor › More features of the framework: h. Databases can also be local files or automatically fetched via ftp or http and also in either FASTA or XML format h. Database Indexes can be automatically built using formatdb h. Multiple input files are joined or split as appropriate to fine-tune throughput h. Output can be delivered via ftp www. cs. wisc. edu/condor
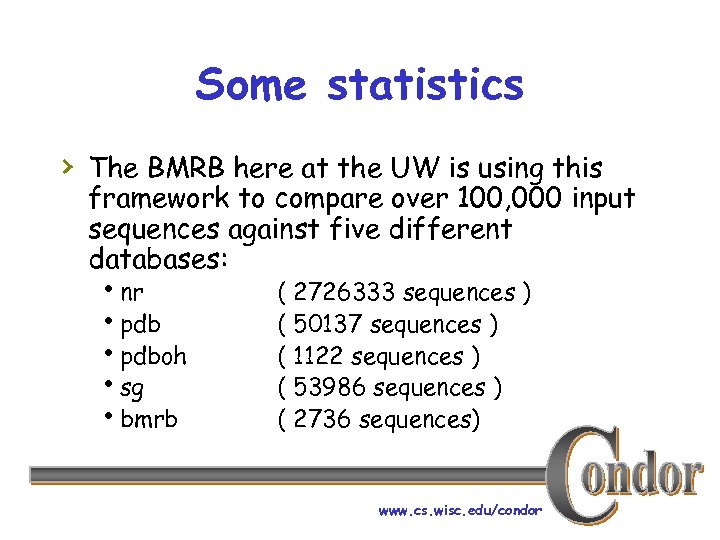 Some statistics › The BMRB here at the UW is using this framework to compare over 100, 000 input sequences against five different databases: hnr hpdboh hsg hbmrb ( 2726333 sequences ) ( 50137 sequences ) ( 1122 sequences ) ( 53986 sequences ) ( 2736 sequences) www. cs. wisc. edu/condor
Some statistics › The BMRB here at the UW is using this framework to compare over 100, 000 input sequences against five different databases: hnr hpdboh hsg hbmrb ( 2726333 sequences ) ( 50137 sequences ) ( 1122 sequences ) ( 53986 sequences ) ( 2736 sequences) www. cs. wisc. edu/condor
 Some statistics › All in all, the BMRB is doing over 8 billion › › sequence comparisons for their weekly run. Condor completes this in roughly eight hours of wall-clock time. This is now a weekly routine which is fully automated, very reliable, and requires almost no “babysitting”. www. cs. wisc. edu/condor
Some statistics › All in all, the BMRB is doing over 8 billion › › sequence comparisons for their weekly run. Condor completes this in roughly eight hours of wall-clock time. This is now a weekly routine which is fully automated, very reliable, and requires almost no “babysitting”. www. cs. wisc. edu/condor
 Structure Calculation › CNS h. Available from http: //cns. csb. yale. edu/ › CYANA h. Available from http: //www. guentert. com/ › Both do structure calculations but use different methods www. cs. wisc. edu/condor
Structure Calculation › CNS h. Available from http: //cns. csb. yale. edu/ › CYANA h. Available from http: //www. guentert. com/ › Both do structure calculations but use different methods www. cs. wisc. edu/condor
 CNS and Condor › Using CNS can take a relatively long time › › to compute for a given entry (protein sequence) depending on the number of possible intermediate structures. Each structure takes about 5 – 30 minutes depending on length of sequence At 200 structures per entry, this ends up being between 16 and 100 hours. www. cs. wisc. edu/condor
CNS and Condor › Using CNS can take a relatively long time › › to compute for a given entry (protein sequence) depending on the number of possible intermediate structures. Each structure takes about 5 – 30 minutes depending on length of sequence At 200 structures per entry, this ends up being between 16 and 100 hours. www. cs. wisc. edu/condor
 CYANA › Cyana takes only about 2 – 16 hours › per entry depending on the sequence length. The cyana results are post-processed with CNS to refine them, which takes an additional 4 – 20 hours per entry www. cs. wisc. edu/condor
CYANA › Cyana takes only about 2 – 16 hours › per entry depending on the sequence length. The cyana results are post-processed with CNS to refine them, which takes an additional 4 – 20 hours per entry www. cs. wisc. edu/condor
 CNS, CYANA, and Condor › Until now, each different group doing › structure calculations would process their own entries using different programs or input parameters, making comparisons between different groups difficult. By processing large numbers of entries in exactly the same way, it is possible to then compare apples to apples. www. cs. wisc. edu/condor
CNS, CYANA, and Condor › Until now, each different group doing › structure calculations would process their own entries using different programs or input parameters, making comparisons between different groups difficult. By processing large numbers of entries in exactly the same way, it is possible to then compare apples to apples. www. cs. wisc. edu/condor
 CNS, CYANA, and Condor › Working with the BMRB, I created a › framework which allows you to easily process multiple entries at once with both CNS and CYANA. Using this framework, Condor calculated structures for 600 entries (about 50, 000 hours) in just 10 days. www. cs. wisc. edu/condor
CNS, CYANA, and Condor › Working with the BMRB, I created a › framework which allows you to easily process multiple entries at once with both CNS and CYANA. Using this framework, Condor calculated structures for 600 entries (about 50, 000 hours) in just 10 days. www. cs. wisc. edu/condor
 CNS, CYANA, and Condor › The structure calculation framework › is also very reliable and requires very little human time to do a fairly massive amount of computing. This process can now be easily automated and done on a routine basis. www. cs. wisc. edu/condor
CNS, CYANA, and Condor › The structure calculation framework › is also very reliable and requires very little human time to do a fairly massive amount of computing. This process can now be easily automated and done on a routine basis. www. cs. wisc. edu/condor
 Challenges › Creating a job flow that doesn’t need › babysitting requires that the framework be able to handle a variety of problems. To this end, it employs some other Condor technologies: h. Many things are wrapped in ftsh. h. Condor watches for “misbehaving” jobs and kills them using the PERIODIC_REMOVE feature. h. DAGMan oversees the whole run and retries failed jobs. www. cs. wisc. edu/condor
Challenges › Creating a job flow that doesn’t need › babysitting requires that the framework be able to handle a variety of problems. To this end, it employs some other Condor technologies: h. Many things are wrapped in ftsh. h. Condor watches for “misbehaving” jobs and kills them using the PERIODIC_REMOVE feature. h. DAGMan oversees the whole run and retries failed jobs. www. cs. wisc. edu/condor
 Future Work › BLAST h. Use STORK for data transfer which will improve reliability of all file transfers and instantly add support for many more methods of transferring input and output. h. Create a wrapper around the framework which behaves just like NCBI’s BLAST but uses Condor behind the scenes. h. Include this framework with the Condor distribution so it is BLAST-ready “out of the box”. www. cs. wisc. edu/condor
Future Work › BLAST h. Use STORK for data transfer which will improve reliability of all file transfers and instantly add support for many more methods of transferring input and output. h. Create a wrapper around the framework which behaves just like NCBI’s BLAST but uses Condor behind the scenes. h. Include this framework with the Condor distribution so it is BLAST-ready “out of the box”. www. cs. wisc. edu/condor
 Future Work › CNS & CYANA h. Use sequence length to better estimate runtime for fine-tuning throughput. h. Use STORK for file transfer. www. cs. wisc. edu/condor
Future Work › CNS & CYANA h. Use sequence length to better estimate runtime for fine-tuning throughput. h. Use STORK for file transfer. www. cs. wisc. edu/condor
 Conclusion › I have created tools which allow users to › › run coordinated BLAST, CNS, and CYANA runs on very large scales. This makes it easy to process not only your data but other groups’ too, and end up with results that were all computed with the same protocols and inputs. This will enable better collaboration by providing more consistency between the results of different groups. www. cs. wisc. edu/condor
Conclusion › I have created tools which allow users to › › run coordinated BLAST, CNS, and CYANA runs on very large scales. This makes it easy to process not only your data but other groups’ too, and end up with results that were all computed with the same protocols and inputs. This will enable better collaboration by providing more consistency between the results of different groups. www. cs. wisc. edu/condor


