0fdb00c46b8b2386eaa493c6416ee4f6.ppt
- Количество слайдов: 23
 Supporting Phenotyping through Visualization and Image Analysis Raghu Machiraju, Computer Science & Engineering, Bio-Medical Informatics The Ohio State University
Supporting Phenotyping through Visualization and Image Analysis Raghu Machiraju, Computer Science & Engineering, Bio-Medical Informatics The Ohio State University
 About Myself v Associate Professor, Computer Science and Engineeering, Bio. Medical Informatics v 7 th Year at OSU v Research Interests – Imaging, Graphics and Visualization v Notable Points v Co-Chair of Visualization 2008 Conference, Columbus OH v Alumni in video gaming/animation industry (Pixar, EA), National Government Labs (Lawrence Livermore), Industrial Research (Samsung, IBM, Mitsubishi Electric), Medical Schools (Harvard Medical School)
About Myself v Associate Professor, Computer Science and Engineeering, Bio. Medical Informatics v 7 th Year at OSU v Research Interests – Imaging, Graphics and Visualization v Notable Points v Co-Chair of Visualization 2008 Conference, Columbus OH v Alumni in video gaming/animation industry (Pixar, EA), National Government Labs (Lawrence Livermore), Industrial Research (Samsung, IBM, Mitsubishi Electric), Medical Schools (Harvard Medical School)
 Research Activities Medical, Biological Imaging and Visualization Ø Optical Microscopy ü In-vivo, fluorescence imaging ü Structural/Functional Magnetic Resonance Imaging ü Diffusion Tensor Imaging • Mostly interested in: • Segmentation, Registration, Tracking • Applications: phenotyping, longitudinal studies
Research Activities Medical, Biological Imaging and Visualization Ø Optical Microscopy ü In-vivo, fluorescence imaging ü Structural/Functional Magnetic Resonance Imaging ü Diffusion Tensor Imaging • Mostly interested in: • Segmentation, Registration, Tracking • Applications: phenotyping, longitudinal studies
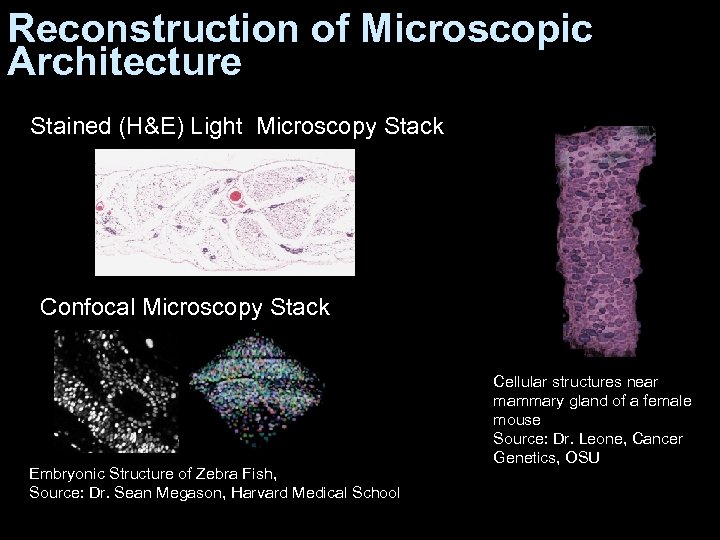 Reconstruction of Microscopic Architecture Stained (H&E) Light Microscopy Stack Confocal Microscopy Stack Embryonic Structure of Zebra Fish, Source: Dr. Sean Megason, Harvard Medical School Cellular structures near mammary gland of a female mouse Source: Dr. Leone, Cancer Genetics, OSU
Reconstruction of Microscopic Architecture Stained (H&E) Light Microscopy Stack Confocal Microscopy Stack Embryonic Structure of Zebra Fish, Source: Dr. Sean Megason, Harvard Medical School Cellular structures near mammary gland of a female mouse Source: Dr. Leone, Cancer Genetics, OSU
 My Colleagues … Kishore Mosaliganti, 5 th year Bioinformatics/Cancer Genetics Gustavo Leone, Mike Ostrowski Human Cancer Genetics Program Kun Huang, Biomedical Informatics
My Colleagues … Kishore Mosaliganti, 5 th year Bioinformatics/Cancer Genetics Gustavo Leone, Mike Ostrowski Human Cancer Genetics Program Kun Huang, Biomedical Informatics
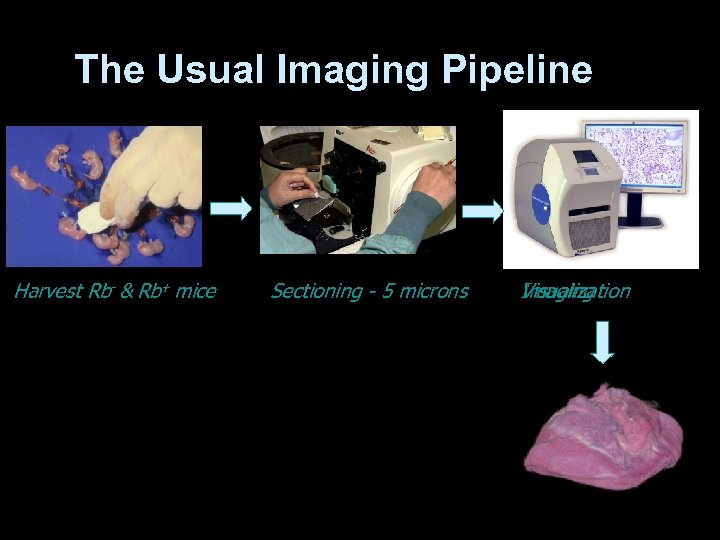 The Usual Imaging Pipeline Harvest Rb- & Rb+ mice Sectioning - 5 microns Visualization Imaging
The Usual Imaging Pipeline Harvest Rb- & Rb+ mice Sectioning - 5 microns Visualization Imaging
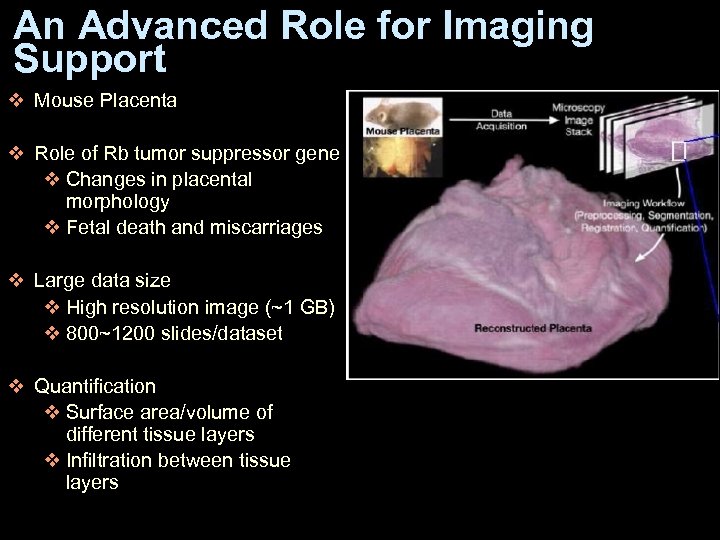 An Advanced Role for Imaging Support v Mouse Placenta v Role of Rb tumor suppressor gene v Changes in placental morphology v Fetal death and miscarriages v Large data size v High resolution image (~1 GB) v 800~1200 slides/dataset v Quantification v Surface area/volume of different tissue layers v Infiltration between tissue layers
An Advanced Role for Imaging Support v Mouse Placenta v Role of Rb tumor suppressor gene v Changes in placental morphology v Fetal death and miscarriages v Large data size v High resolution image (~1 GB) v 800~1200 slides/dataset v Quantification v Surface area/volume of different tissue layers v Infiltration between tissue layers
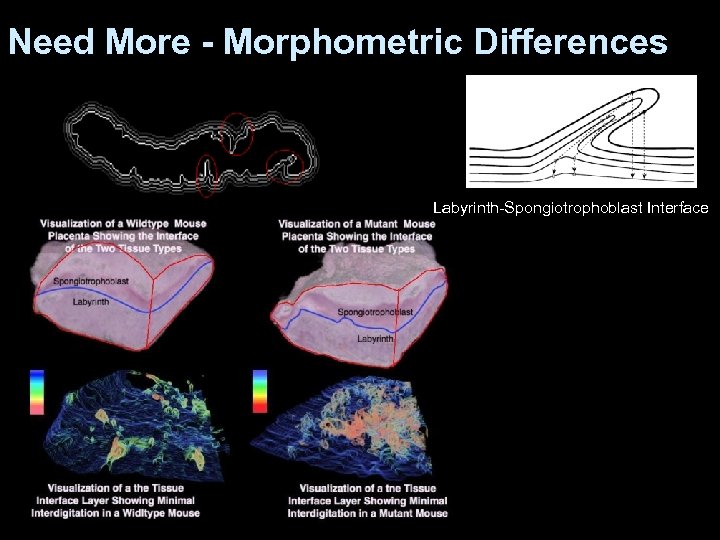 Need More - Morphometric Differences Labyrinth-Spongiotrophoblast Interface
Need More - Morphometric Differences Labyrinth-Spongiotrophoblast Interface
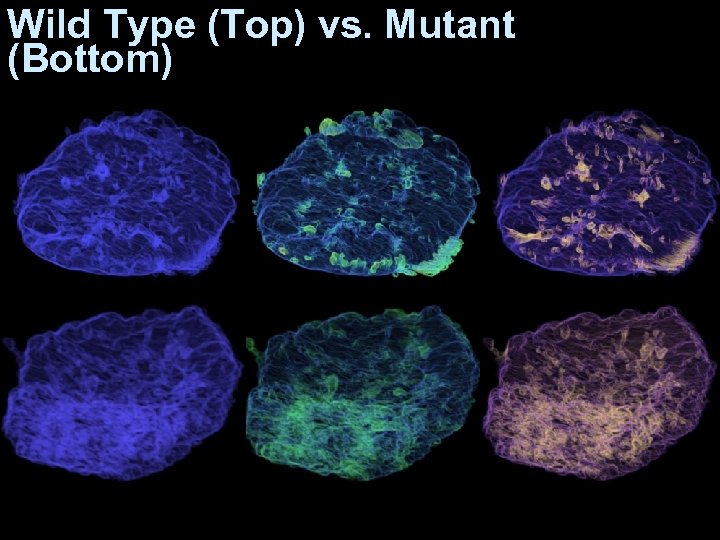 Wild Type (Top) vs. Mutant (Bottom)
Wild Type (Top) vs. Mutant (Bottom)
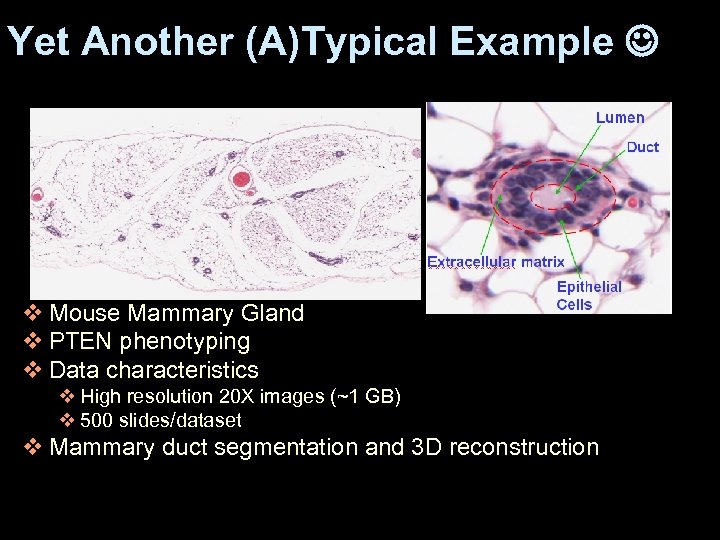 Yet Another (A)Typical Example v Mouse Mammary Gland v PTEN phenotyping v Data characteristics v High resolution 20 X images (~1 GB) v 500 slides/dataset v Mammary duct segmentation and 3 D reconstruction
Yet Another (A)Typical Example v Mouse Mammary Gland v PTEN phenotyping v Data characteristics v High resolution 20 X images (~1 GB) v 500 slides/dataset v Mammary duct segmentation and 3 D reconstruction
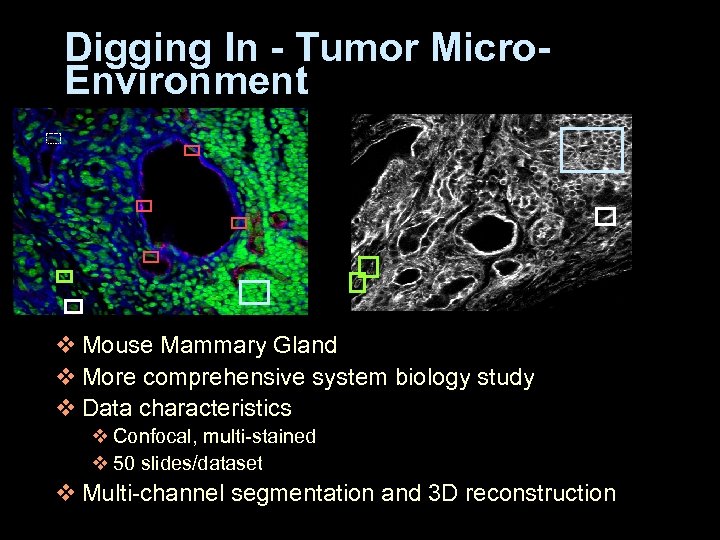 Digging In - Tumor Micro. Environment v Mouse Mammary Gland v More comprehensive system biology study v Data characteristics v Confocal, multi-stained v 50 slides/dataset v Multi-channel segmentation and 3 D reconstruction
Digging In - Tumor Micro. Environment v Mouse Mammary Gland v More comprehensive system biology study v Data characteristics v Confocal, multi-stained v 50 slides/dataset v Multi-channel segmentation and 3 D reconstruction
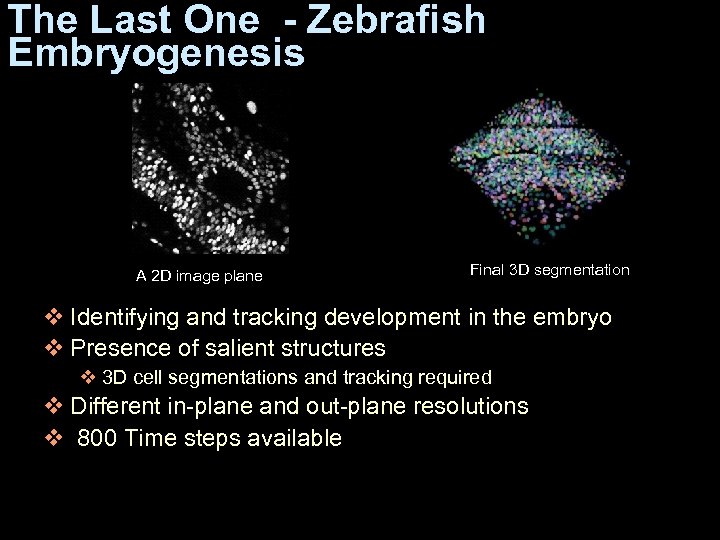 The Last One - Zebrafish Embryogenesis A 2 D image plane Final 3 D segmentation v Identifying and tracking development in the embryo v Presence of salient structures v 3 D cell segmentations and tracking required v Different in-plane and out-plane resolutions v 800 Time steps available
The Last One - Zebrafish Embryogenesis A 2 D image plane Final 3 D segmentation v Identifying and tracking development in the embryo v Presence of salient structures v 3 D cell segmentations and tracking required v Different in-plane and out-plane resolutions v 800 Time steps available
 The Underlying Premise Is there an unified way to visualize and analyze the various microscopic image modalities ?
The Underlying Premise Is there an unified way to visualize and analyze the various microscopic image modalities ?
 The Essentials Of Microstructure v Premise - you can measure, visualize and analyze cellular structures if you characterize and build virtual microstructure v Component v. Distributions v. Packing v. Arrangements v Material Interfaces
The Essentials Of Microstructure v Premise - you can measure, visualize and analyze cellular structures if you characterize and build virtual microstructure v Component v. Distributions v. Packing v. Arrangements v Material Interfaces
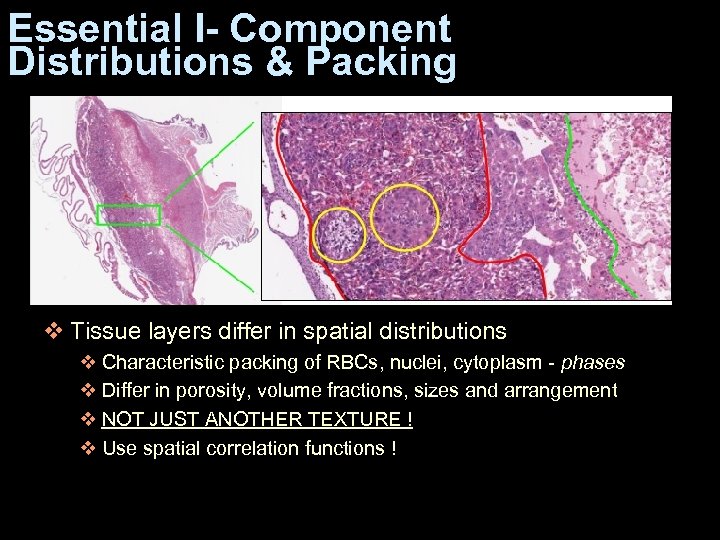 Essential I- Component Distributions & Packing v Tissue layers differ in spatial distributions v Characteristic packing of RBCs, nuclei, cytoplasm - phases v Differ in porosity, volume fractions, sizes and arrangement v NOT JUST ANOTHER TEXTURE ! v Use spatial correlation functions !
Essential I- Component Distributions & Packing v Tissue layers differ in spatial distributions v Characteristic packing of RBCs, nuclei, cytoplasm - phases v Differ in porosity, volume fractions, sizes and arrangement v NOT JUST ANOTHER TEXTURE ! v Use spatial correlation functions !
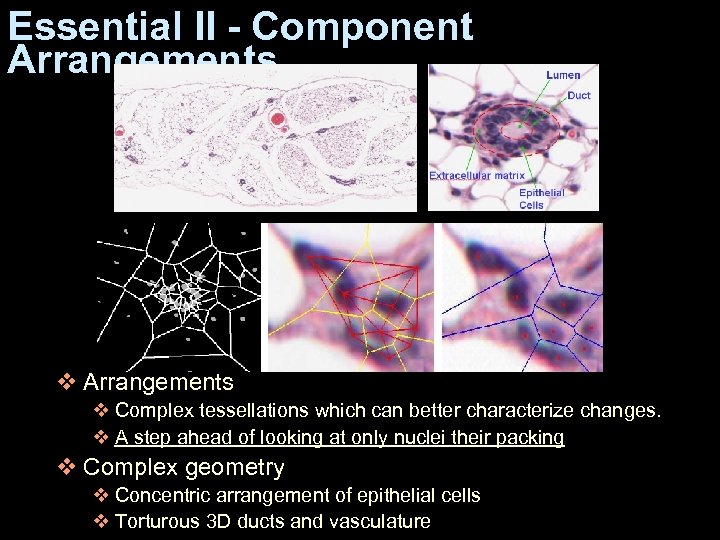 Essential II - Component Arrangements v Complex tessellations which can better characterize changes. v A step ahead of looking at only nuclei their packing v Complex geometry v Concentric arrangement of epithelial cells v Torturous 3 D ducts and vasculature
Essential II - Component Arrangements v Complex tessellations which can better characterize changes. v A step ahead of looking at only nuclei their packing v Complex geometry v Concentric arrangement of epithelial cells v Torturous 3 D ducts and vasculature
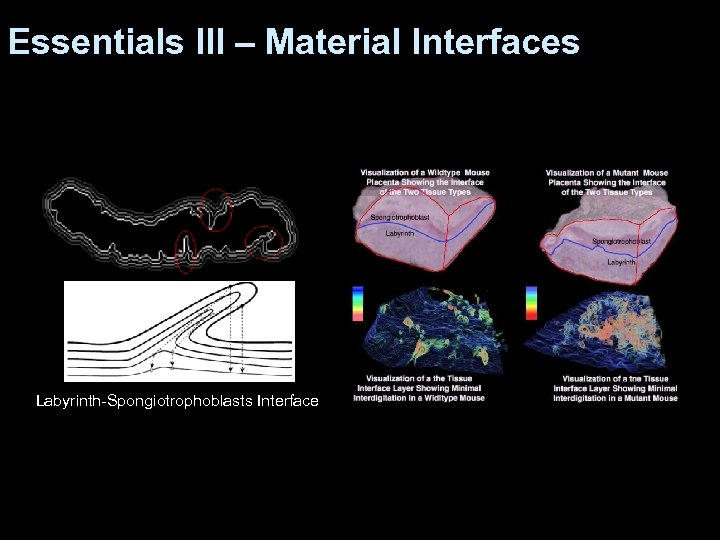 Essentials III – Material Interfaces Labyrinth-Spongiotrophoblasts Interface
Essentials III – Material Interfaces Labyrinth-Spongiotrophoblasts Interface
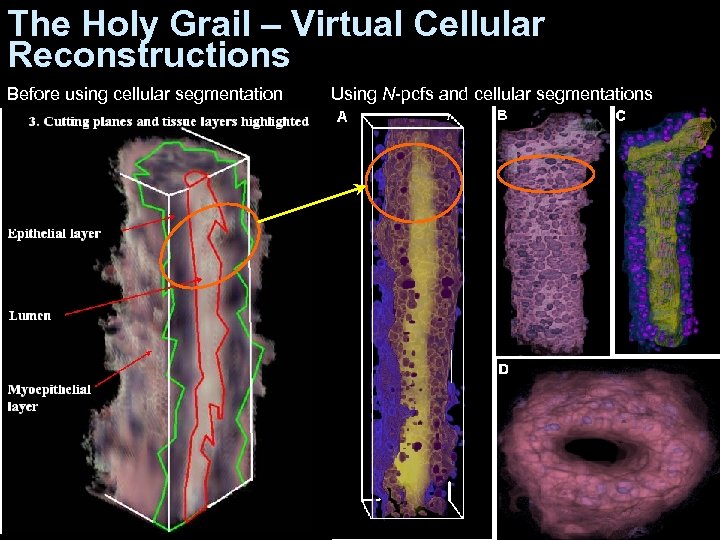 The Holy Grail – Virtual Cellular Reconstructions Before using cellular segmentation Using N-pcfs and cellular segmentations
The Holy Grail – Virtual Cellular Reconstructions Before using cellular segmentation Using N-pcfs and cellular segmentations
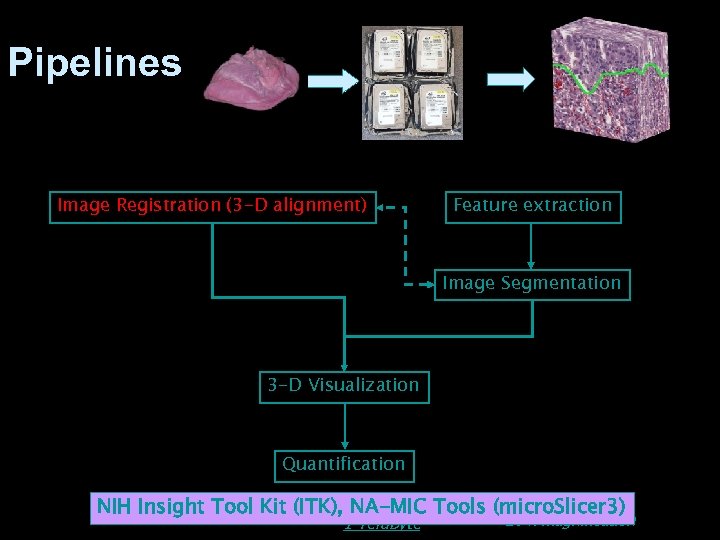 Pipelines Image Registration (3 -D alignment) Feature extraction Image Segmentation 3 -D Visualization Quantification 1 Gb x 1 Gb x 900 NIH Insight Tool Kit (ITK), NA-MIC Tools (micro. Slicer 3) 1 Tera. Byte 20 x magnification
Pipelines Image Registration (3 -D alignment) Feature extraction Image Segmentation 3 -D Visualization Quantification 1 Gb x 1 Gb x 900 NIH Insight Tool Kit (ITK), NA-MIC Tools (micro. Slicer 3) 1 Tera. Byte 20 x magnification
 Conclusions v Highly multi-disciplinary approach. v Need scalability and robustness v Useful workflows need to be constructed v Much application-domain knowledge has to be embedded in algorithms v Validation of methods and proving robustness is a pre-occupation. v The final goal of a virtual cellular architecture is not that elusive
Conclusions v Highly multi-disciplinary approach. v Need scalability and robustness v Useful workflows need to be constructed v Much application-domain knowledge has to be embedded in algorithms v Validation of methods and proving robustness is a pre-occupation. v The final goal of a virtual cellular architecture is not that elusive
 Destroying The Amazon Rain Forest v K. Mosaliganti and R. Machiraju et al. An Imaging Workflow for Characterizing Phenotypical Change in Terabyte Sized Mouse Model Datasets. Journal of Bioinformatics, 2008 (to appear) v K. Mosaliganti and R. Machiraju et al. Visualization of Cellular Biology Structures from Optical Microscopy Data. IEEE Transactions in Visualization and Computer Graphics, 2008 (to appear) v K. Mosaliganti, R. Machiraju et al. Tensor Classification of N-point Correlation Function features for Histology Tissue Segmentation. Journal of Medical Image Analysis, 2008 (to appear) v K. Mosaliganti and R. Machiraju et al. Geometry-driven Visualization of Microscopic Structures in Biology. Workshop on Knowledge-Assisted Visualization, Proceedings of Euro. Vis 2008 (to appear). v K. Mosaliganti, R. Machiraju et al. “Detection and Visualization of Surface-Pockets to Enable Phenotyping Studies”. IEEE Transactions on Medical Imaging, volume 26(9), pages 1283 -1290, 2007. v R. Sharp, K. Mosaliganti et al. “Volume Rendering Phenotype Differences in Mouse Placenta Microscopy Data”. Journal of Computing in Science and Engineering, volume 9 (1), pages 38 -47, Jan/ Feb 2007. v P. Wenzel and K. Mosaliganti et al. Rb is critical in a mammalian tissue stem cell population. In Journal of Genetics and Development, volume 21 (1), pages 85 -97, Jan 2007. v K. Mosaliganti and R. Machiraju et al. Automated Quantification of Colony Growth in Clonogenic Assays. Workshop on Medical Image Analysis with Applications in Biology, 2007, Piscatway, Rutgers, New Jersey, USA. v R. Ridgway, R. Machiraju et al. Image segmentation with tensor-based classification of N-point correlation functions. In MICCAI Workshop on Medical Image Analysis with Applications in Biology, 2006. v O. Irfanoglu, K. Mosaliganti et al. “Histology Image Segmentation using the N-Point Correlation Functions”. International Symposium of Biomedical Imaging, 2006. v
Destroying The Amazon Rain Forest v K. Mosaliganti and R. Machiraju et al. An Imaging Workflow for Characterizing Phenotypical Change in Terabyte Sized Mouse Model Datasets. Journal of Bioinformatics, 2008 (to appear) v K. Mosaliganti and R. Machiraju et al. Visualization of Cellular Biology Structures from Optical Microscopy Data. IEEE Transactions in Visualization and Computer Graphics, 2008 (to appear) v K. Mosaliganti, R. Machiraju et al. Tensor Classification of N-point Correlation Function features for Histology Tissue Segmentation. Journal of Medical Image Analysis, 2008 (to appear) v K. Mosaliganti and R. Machiraju et al. Geometry-driven Visualization of Microscopic Structures in Biology. Workshop on Knowledge-Assisted Visualization, Proceedings of Euro. Vis 2008 (to appear). v K. Mosaliganti, R. Machiraju et al. “Detection and Visualization of Surface-Pockets to Enable Phenotyping Studies”. IEEE Transactions on Medical Imaging, volume 26(9), pages 1283 -1290, 2007. v R. Sharp, K. Mosaliganti et al. “Volume Rendering Phenotype Differences in Mouse Placenta Microscopy Data”. Journal of Computing in Science and Engineering, volume 9 (1), pages 38 -47, Jan/ Feb 2007. v P. Wenzel and K. Mosaliganti et al. Rb is critical in a mammalian tissue stem cell population. In Journal of Genetics and Development, volume 21 (1), pages 85 -97, Jan 2007. v K. Mosaliganti and R. Machiraju et al. Automated Quantification of Colony Growth in Clonogenic Assays. Workshop on Medical Image Analysis with Applications in Biology, 2007, Piscatway, Rutgers, New Jersey, USA. v R. Ridgway, R. Machiraju et al. Image segmentation with tensor-based classification of N-point correlation functions. In MICCAI Workshop on Medical Image Analysis with Applications in Biology, 2006. v O. Irfanoglu, K. Mosaliganti et al. “Histology Image Segmentation using the N-Point Correlation Functions”. International Symposium of Biomedical Imaging, 2006. v
 Acknowledgements v Joel Saltz, BMI v Richard Sharp, Okan Irfanoglu, Firdaus Janoos, CSE OSU v Weiming Xia, Sean Megason, Harvard Medical school v Jens Rittscher, GE Global Research v NIH, NLM Training Grant v NSF ITR grant
Acknowledgements v Joel Saltz, BMI v Richard Sharp, Okan Irfanoglu, Firdaus Janoos, CSE OSU v Weiming Xia, Sean Megason, Harvard Medical school v Jens Rittscher, GE Global Research v NIH, NLM Training Grant v NSF ITR grant
 Thank You ! Questions ?
Thank You ! Questions ?


