f7abe2e64c6c4d01f4d6364d705c6b16.ppt
- Количество слайдов: 17
 Suicide Polymerase Endonuclease Restriction (Su. PER) PCR: A NOVEL MOLECULAR MICROBIOLOGICAL METHOD FOR DETECTING THE HARD TO DETECT Stefan J. Green 1, 2 and Dror Minz 2 1. Faculty of Agricultural, Food and Environmental Quality Sciences, Hebrew University of Jerusalem, Rehovot 2. The Volcani Institute, Agricultural Research Organization, Bet-Dagan, Israel 10 th International Symposium on Microbial Ecology ISME-10 Cancun, Mexico, Monday, August 23 rd, 2004 Green, S. J. and Minz, D. 2005. Suicide Polymerase Endonuclease Restriction (Su. PER) – a novel technique for enhancing PCR amplification of minor DNA templates. Appl. Environ. Microbiol. 71: 4721 -4727. http: //www. superpcr. blogspot. com/
Suicide Polymerase Endonuclease Restriction (Su. PER) PCR: A NOVEL MOLECULAR MICROBIOLOGICAL METHOD FOR DETECTING THE HARD TO DETECT Stefan J. Green 1, 2 and Dror Minz 2 1. Faculty of Agricultural, Food and Environmental Quality Sciences, Hebrew University of Jerusalem, Rehovot 2. The Volcani Institute, Agricultural Research Organization, Bet-Dagan, Israel 10 th International Symposium on Microbial Ecology ISME-10 Cancun, Mexico, Monday, August 23 rd, 2004 Green, S. J. and Minz, D. 2005. Suicide Polymerase Endonuclease Restriction (Su. PER) – a novel technique for enhancing PCR amplification of minor DNA templates. Appl. Environ. Microbiol. 71: 4721 -4727. http: //www. superpcr. blogspot. com/
 Su. PER PCR What is Su. PER PCR? • A molecular method to selectively destroy, via endonuclease restriction, target DNA templates.
Su. PER PCR What is Su. PER PCR? • A molecular method to selectively destroy, via endonuclease restriction, target DNA templates.
 Su. PER PCR How does it work? • The Su. PER reaction relies on two basic premises: • PCR primers can bind stringently to target DNA templates • Restriction enzymes digest double-stranded DNA only.
Su. PER PCR How does it work? • The Su. PER reaction relies on two basic premises: • PCR primers can bind stringently to target DNA templates • Restriction enzymes digest double-stranded DNA only.
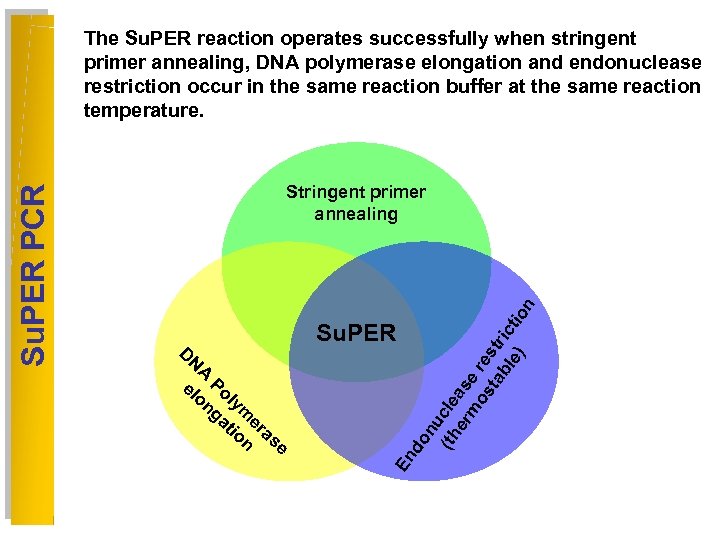 Su. PER D N A el Po on ly ga me tio ra n s e do nu (th cle er as m er os e ta str bl ic e) tio n Stringent primer annealing En Su. PER PCR The Su. PER reaction operates successfully when stringent primer annealing, DNA polymerase elongation and endonuclease restriction occur in the same reaction buffer at the same reaction temperature.
Su. PER D N A el Po on ly ga me tio ra n s e do nu (th cle er as m er os e ta str bl ic e) tio n Stringent primer annealing En Su. PER PCR The Su. PER reaction operates successfully when stringent primer annealing, DNA polymerase elongation and endonuclease restriction occur in the same reaction buffer at the same reaction temperature.
 Su. PER PCR STEP 1: Double-stranded DNA is denatured.
Su. PER PCR STEP 1: Double-stranded DNA is denatured.
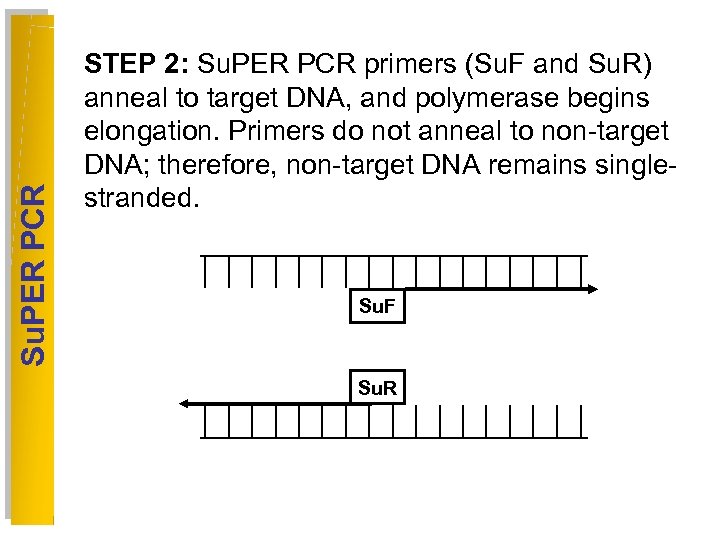 Su. PER PCR STEP 2: Su. PER PCR primers (Su. F and Su. R) anneal to target DNA, and polymerase begins elongation. Primers do not anneal to non-target DNA; therefore, non-target DNA remains singlestranded. Su. F Su. R
Su. PER PCR STEP 2: Su. PER PCR primers (Su. F and Su. R) anneal to target DNA, and polymerase begins elongation. Primers do not anneal to non-target DNA; therefore, non-target DNA remains singlestranded. Su. F Su. R
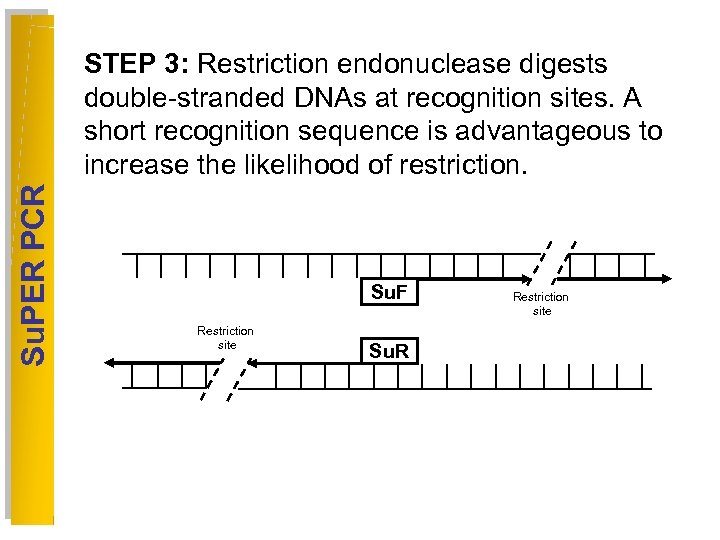 Su. PER PCR STEP 3: Restriction endonuclease digests double-stranded DNAs at recognition sites. A short recognition sequence is advantageous to increase the likelihood of restriction. Su. F Restriction site Su. R Restriction site
Su. PER PCR STEP 3: Restriction endonuclease digests double-stranded DNAs at recognition sites. A short recognition sequence is advantageous to increase the likelihood of restriction. Su. F Restriction site Su. R Restriction site
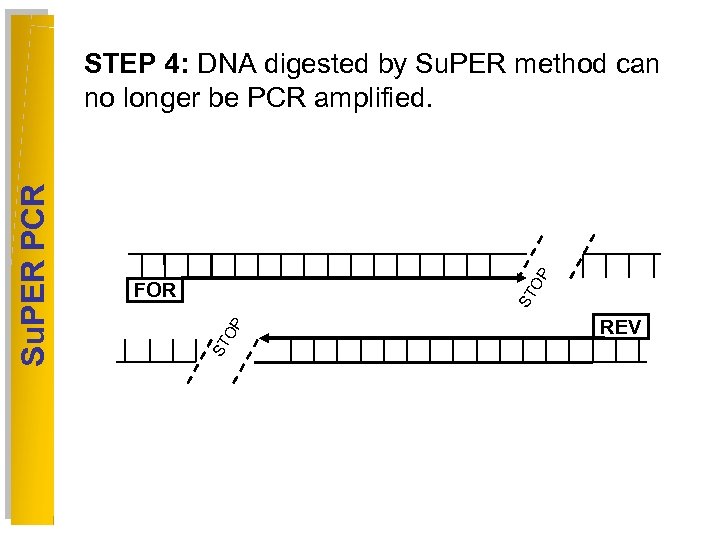 OP OP ST FOR ST Su. PER PCR STEP 4: DNA digested by Su. PER method can no longer be PCR amplified. REV
OP OP ST FOR ST Su. PER PCR STEP 4: DNA digested by Su. PER method can no longer be PCR amplified. REV
 Demonstration: Su. PER PCR Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples PCR-DGGE analysis employing general bacterial primers amplifying an approximately 500 bp fragment of the 16 S r. DNA from the region 341 to 926 (E. coli numbering)
Demonstration: Su. PER PCR Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples PCR-DGGE analysis employing general bacterial primers amplifying an approximately 500 bp fragment of the 16 S r. DNA from the region 341 to 926 (E. coli numbering)
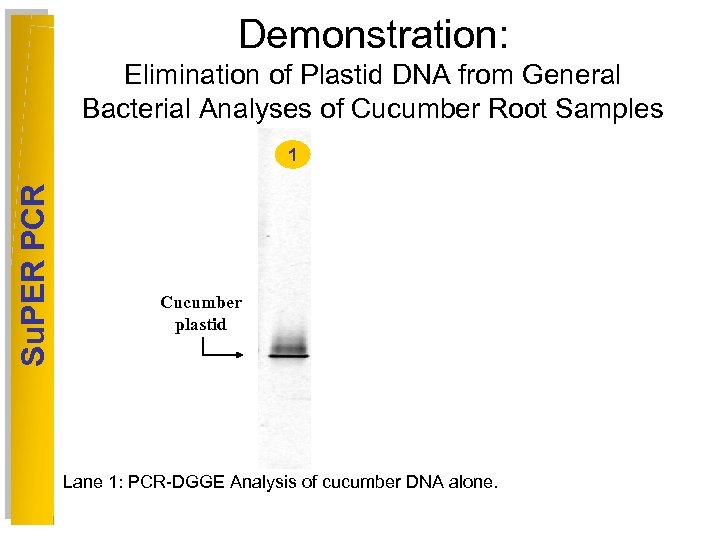 Demonstration: Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples Su. PER PCR 1 2 3 4 Cucumber plastid Lane 1: PCR-DGGE Analysis of cucumber DNA alone. 5 6
Demonstration: Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples Su. PER PCR 1 2 3 4 Cucumber plastid Lane 1: PCR-DGGE Analysis of cucumber DNA alone. 5 6
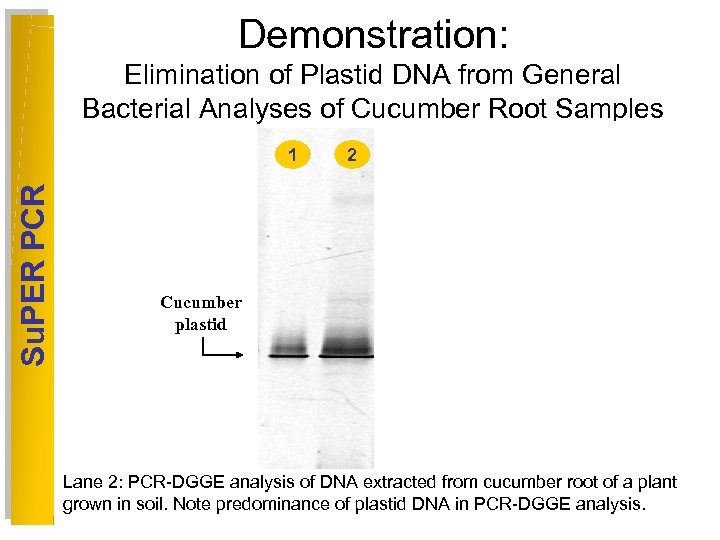 Demonstration: Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples Su. PER PCR 1 2 3 4 5 6 Cucumber plastid Lane 2: PCR-DGGE analysis of DNA extracted from cucumber root of a plant grown in soil. Note predominance of plastid DNA in PCR-DGGE analysis.
Demonstration: Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples Su. PER PCR 1 2 3 4 5 6 Cucumber plastid Lane 2: PCR-DGGE analysis of DNA extracted from cucumber root of a plant grown in soil. Note predominance of plastid DNA in PCR-DGGE analysis.
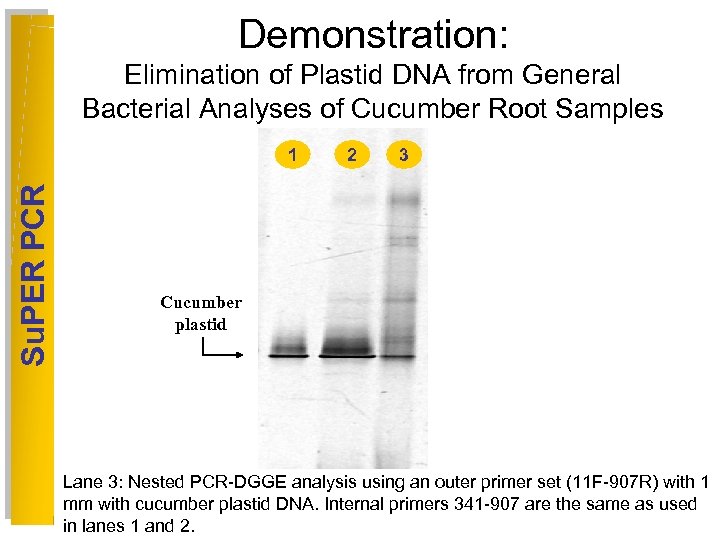 Demonstration: Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples Su. PER PCR 1 2 3 4 5 6 Cucumber plastid Lane 3: Nested PCR-DGGE analysis using an outer primer set (11 F-907 R) with 1 mm with cucumber plastid DNA. Internal primers 341 -907 are the same as used in lanes 1 and 2.
Demonstration: Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples Su. PER PCR 1 2 3 4 5 6 Cucumber plastid Lane 3: Nested PCR-DGGE analysis using an outer primer set (11 F-907 R) with 1 mm with cucumber plastid DNA. Internal primers 341 -907 are the same as used in lanes 1 and 2.
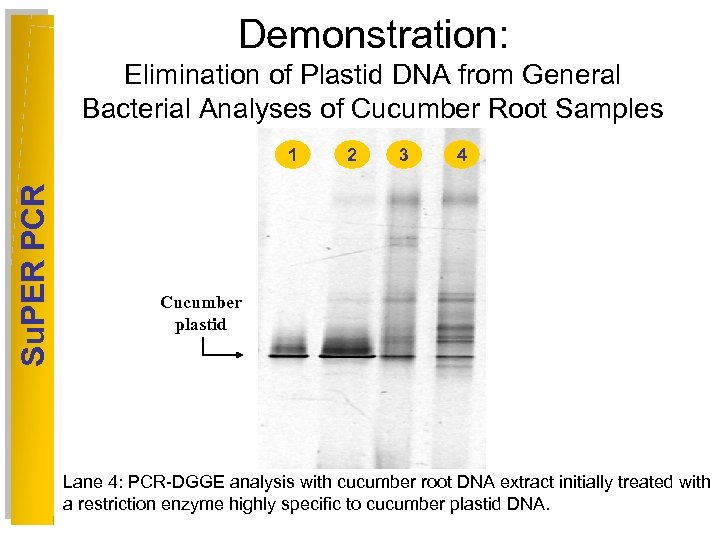 Demonstration: Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples Su. PER PCR 1 2 3 4 5 6 Cucumber plastid Lane 4: PCR-DGGE analysis with cucumber root DNA extract initially treated with a restriction enzyme highly specific to cucumber plastid DNA.
Demonstration: Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples Su. PER PCR 1 2 3 4 5 6 Cucumber plastid Lane 4: PCR-DGGE analysis with cucumber root DNA extract initially treated with a restriction enzyme highly specific to cucumber plastid DNA.
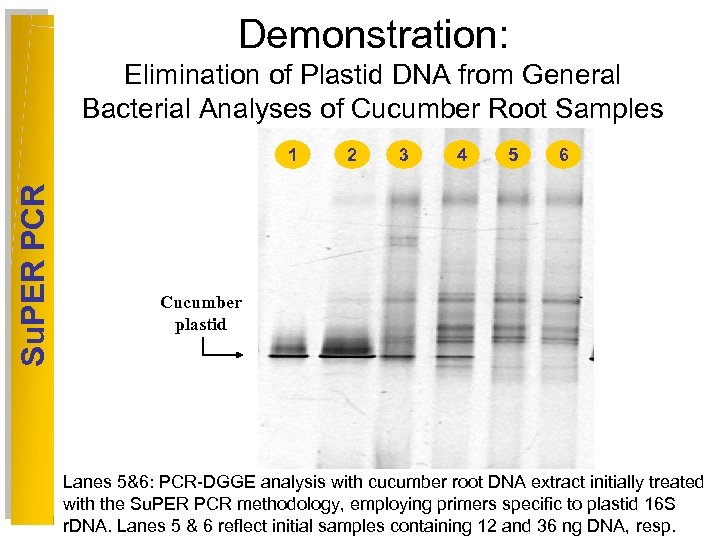 Demonstration: Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples Su. PER PCR 1 2 3 4 5 6 Cucumber plastid Lanes 5&6: PCR-DGGE analysis with cucumber root DNA extract initially treated with the Su. PER PCR methodology, employing primers specific to plastid 16 S r. DNA. Lanes 5 & 6 reflect initial samples containing 12 and 36 ng DNA, resp.
Demonstration: Elimination of Plastid DNA from General Bacterial Analyses of Cucumber Root Samples Su. PER PCR 1 2 3 4 5 6 Cucumber plastid Lanes 5&6: PCR-DGGE analysis with cucumber root DNA extract initially treated with the Su. PER PCR methodology, employing primers specific to plastid 16 S r. DNA. Lanes 5 & 6 reflect initial samples containing 12 and 36 ng DNA, resp.
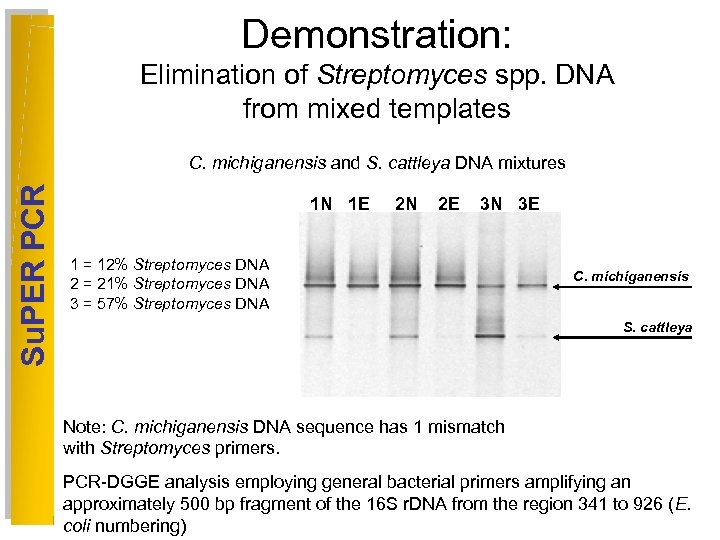 Demonstration: Elimination of Streptomyces spp. DNA from mixed templates Su. PER PCR C. michiganensis and S. cattleya DNA mixtures 1 N 1 E 2 N 2 E 3 N 3 E 1 = 12% Streptomyces DNA 2 = 21% Streptomyces DNA 3 = 57% Streptomyces DNA C. michiganensis S. cattleya Note: C. michiganensis DNA sequence has 1 mismatch with Streptomyces primers. PCR-DGGE analysis employing general bacterial primers amplifying an approximately 500 bp fragment of the 16 S r. DNA from the region 341 to 926 (E. coli numbering)
Demonstration: Elimination of Streptomyces spp. DNA from mixed templates Su. PER PCR C. michiganensis and S. cattleya DNA mixtures 1 N 1 E 2 N 2 E 3 N 3 E 1 = 12% Streptomyces DNA 2 = 21% Streptomyces DNA 3 = 57% Streptomyces DNA C. michiganensis S. cattleya Note: C. michiganensis DNA sequence has 1 mismatch with Streptomyces primers. PCR-DGGE analysis employing general bacterial primers amplifying an approximately 500 bp fragment of the 16 S r. DNA from the region 341 to 926 (E. coli numbering)
 Su. PER PCR Summary • The Su. PER reaction can be used to eliminate a troublesome DNA template prior to PCR amplification with general primers. • The Su. PER reaction may also be employed prior to ligation of DNA template into plasmids, or may be used to eliminate single strands of DNA. • The reaction can be highly stringent and can discriminate against a single mismatch. • This method can be applied to medical, forensic and microbial ecology fields or any other where an undesired DNA type inhibits molecular studies.
Su. PER PCR Summary • The Su. PER reaction can be used to eliminate a troublesome DNA template prior to PCR amplification with general primers. • The Su. PER reaction may also be employed prior to ligation of DNA template into plasmids, or may be used to eliminate single strands of DNA. • The reaction can be highly stringent and can discriminate against a single mismatch. • This method can be applied to medical, forensic and microbial ecology fields or any other where an undesired DNA type inhibits molecular studies.
 A similar concept for RNA • Yutaka Uyeno, Yuji Sekiguchi, Akiko Sunaga, Hiroki Yoshida, and Yoichi Kamagata (2004). Sequence-Specific Cleavage of Small-Subunit (SSU) r. RNA with Oligonucleotides and RNase H: a Rapid and Simple Approach to SSU r. RNABased Quantitative Detection of Microorganisms. Appl. Environ. Microbiol. 70: 3650 -3663.
A similar concept for RNA • Yutaka Uyeno, Yuji Sekiguchi, Akiko Sunaga, Hiroki Yoshida, and Yoichi Kamagata (2004). Sequence-Specific Cleavage of Small-Subunit (SSU) r. RNA with Oligonucleotides and RNase H: a Rapid and Simple Approach to SSU r. RNABased Quantitative Detection of Microorganisms. Appl. Environ. Microbiol. 70: 3650 -3663.


