2f7a53630d68c97e5bb0c8cb98f6e2c1.ppt
- Количество слайдов: 22

“Standardising Analytical Metabonomics”

AUTh bio. Analytical group Metabonomics • Fundamental / Developmental work New Methods (Targeted, Untargeted) New Materials Validation • Clinical Studies Rheumatoid Arthritis Physical Exercise Frailty Embryo. Metabolomics Sepsis/NEC newborns
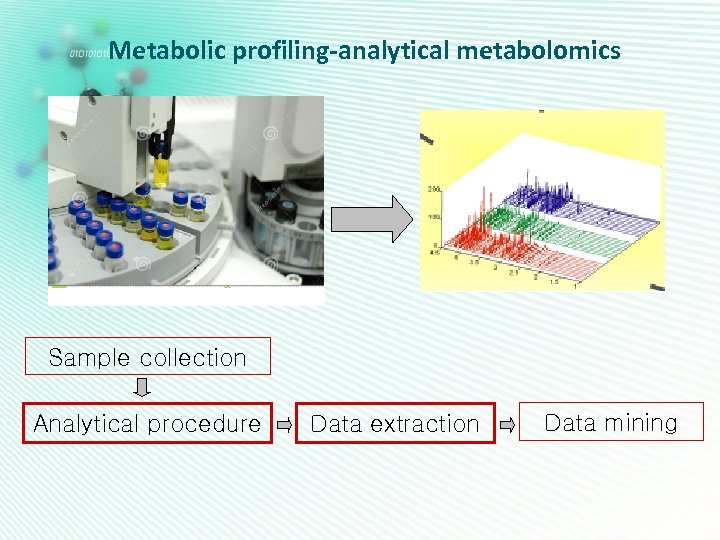
Metabolic profiling-analytical metabolomics Sample collection Analytical procedure Data extraction Data mining
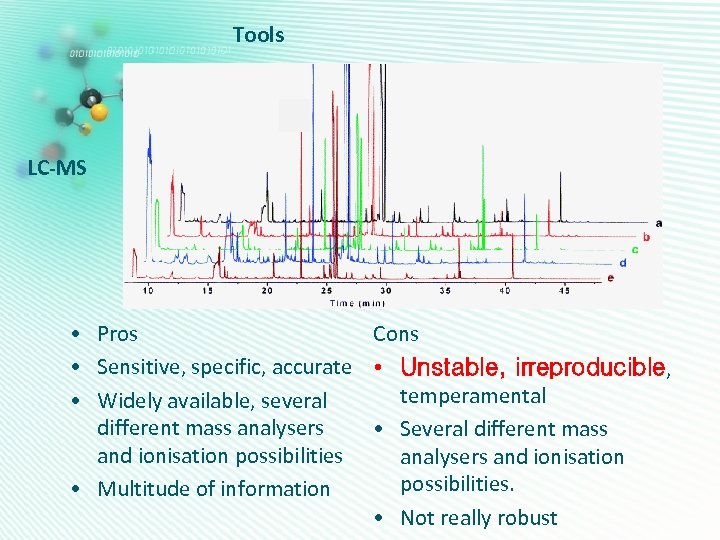
Tools LC-MS Cons • Pros • Sensitive, specific, accurate • Unstable, irreproducible, temperamental • Widely available, several different mass analysers • Several different mass and ionisation possibilities analysers and ionisation possibilities. • Multitude of information • Not really robust
Bottlenecks in analytical procedure • • LC-MS instrumentation variability: Drifts in Rt, mass, sensitivity Need for long analytical batches Unknown trends /unknown components-analytes Instrument calibration along the run Different instrumentations/architecture Wide spectrum of analytes Huge span in concentration: 7 orders of magnitude Full scan mode aqcuisition
Bottlenecks in data treatment • • Big datasets Impractical to correlate-combine data Various picking and treatment algorithms Filtering of noise analytically-oriented lack of data repositories and databases lack of commercial or wide-use LC-MS spectra libraries met. ID • Analytical Chemists, Informaticians, Chemometricians, biochemists still speak different language
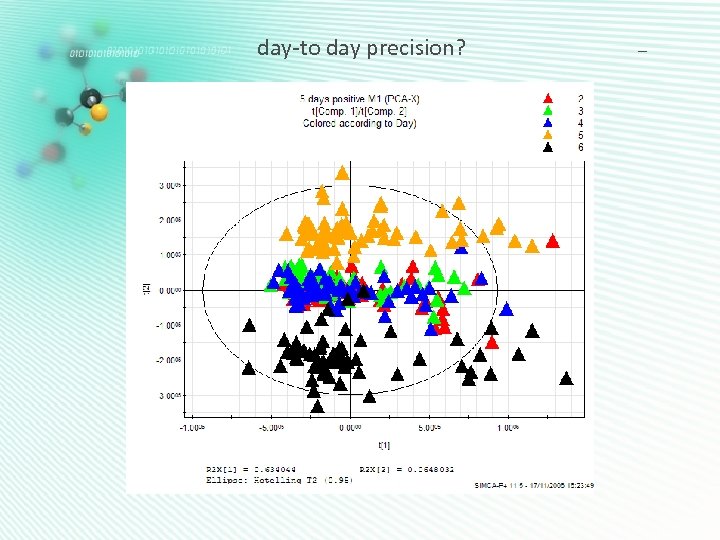
day-to day precision? -

Need for standardization & harmonisation Establishing guidelines/ SOPs • • Data quality (accuracy and precision of measurements) QC procedures Instrument performance and maintenance Sample collection/storage Sample treatment Data acquisition protocols Data manipulation
How can we validate a metabolic profiling method when we don’t know the analytes in advance Integration of classical analytical strategies with modern unbiased data analysis • Implementation of QC (pooled study sample analyzed at regular intervals) • Synthetic mixtures injections • Randomisation of injection order • Technical replicates
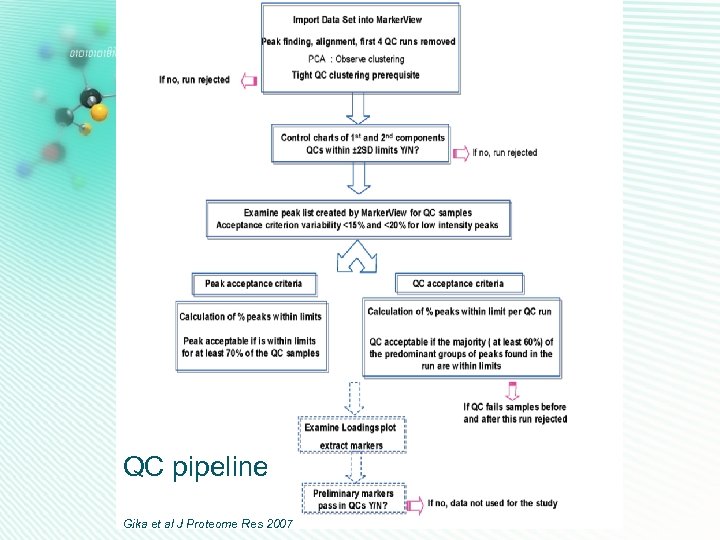
QC pipeline Gika et al J Proteome Res 2007

The project “Standardising Analytical Metabonomics” Co-funded by European social fund and national sources

Scope • To promote standardization and quality control • To address major bottlenecks in analytical practice (development of advanced analytical methodologies for. MS-based metabolic profiling) • To develop informatics tools to improve the quality of the extracted information
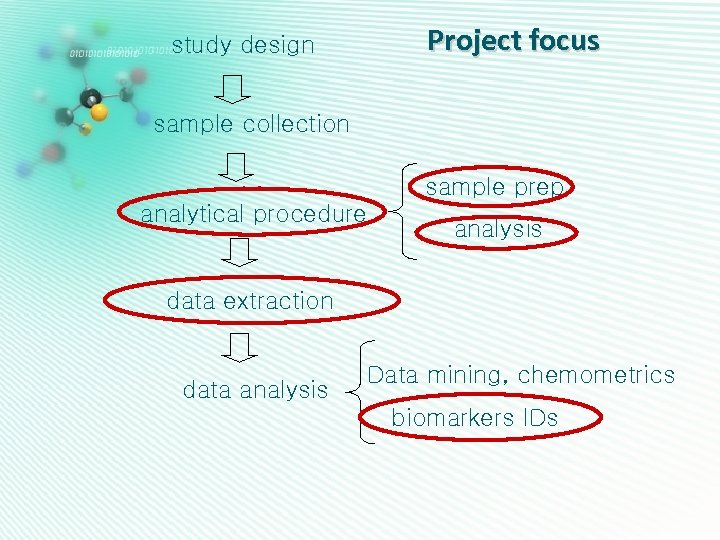
Project focus study design sample collection sample prep analytical procedure analysis data extraction data analysis Data mining, chemometrics biomarkers IDs
WP 1: Development of Analytical Methodologies Method robustness Extraction efficiency Metabolome coverage • Profiling methods with complementary/orthogonal selectivities -HILIC/MS-MS for quantitative determination of ca. 140 primary metabolites -Implementation of other HILIC chemistries eg zwitterionic, diol, RP-WAX - Computational approach for column selection for metabolic profiling • Protocols for sample extraction -Optimization studies on extraction of feces samples, tissue etc (e. g. different p. H values, organic solvent composition, mass to volume ratio) -Liquid and Solid Phase Extraction (SPE) assays for the fractionation of the extract minimising ion suppression effects/compatibility with MS Derivatisation conditions optimization for GC-MS
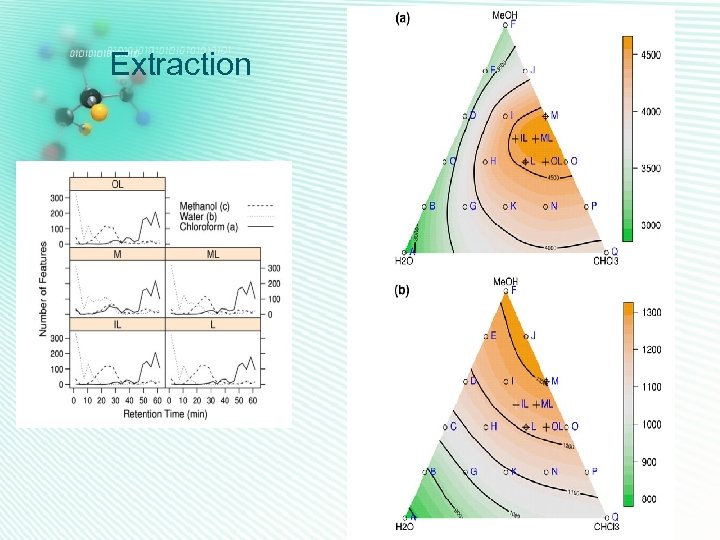
Extraction
WP 2: Data extraction • Evaluation of various data extraction software (free and commercial: XCMS, Marker. Lynx, Marker. View, Profiler and others) in real metabonomics studies. • Spiking experiments (comparison of sensitivity and reliability of the data treatment software) • Development of intranet platform for the extraction of information from MSprofiling data (rules for monitoring and reporting the various alterations and parameter selection to improve standardization in data extraction and reporting

WP 3: Quality Control and standardisation protocols • Scripts for QC in holistic MS data • Examine data in depth and applying rules by automated scripts • Correction for retention time drift to improve peak alignment in feature detection. • Unifying these utilities in one program to standardize promote and consolidate quality control and minimize error possibilities.
WP 4: Data fusion • Software tools to fuse data from different methods LC-MS/MS + GC-MS LC-MS/MS + NMR HILIC-MS + RPLC-MS +evi ESi/ -evi ESI • link data • combine into one table of features or metabolites (? )
WP 5: Metabolite Identification Met. ID the major bottleneck in LC-MS metabonomics • scripts for adduct identification to reduce the number of detected features : +Na+, + NH 4+ , dimers etc • MS spectra by analysis of standards (in-house MS database). • Scripts for automated searches in local and internetbased spectral/biochemistry libraries. • Compare isotope patterns between peaks in samples and standards
WP 6 : Retention Time Prediction • Incorporating Rt data to assists Met. ID • Use of data from orthogonal chromatographic systems: chemical information (polarity, Log. P etc) • Rule out candidate IDs Retention time prediction algorithm in HILIC • software to organise the necessary analyses and data treatment for met. ID within an easy to use platform.

Summary • • Strong need for Standardisation LC-MS is a major part of the solution (and the problem!) Metabolomics is analytically dependent Intelligent tools are needed to go through data and efficiently check data quality The major aim is to find biomarkers – when you’ve found them the real work begins.

The group Auth • Dr. H. Gika • Dr. G. Theodoridis • Prof. A. Papa • Dr. N. Raikos • Dr. C. Zisi • Dr. C. Liambas • O. Deda MSc • S. Fasoula MSc • A. C. Hatzioannou MSc • D. Palachanis MSc • C. Virgiliou MSc • I. Sampsonidis MSc External collaborators • I. D. Wilson Imperial college London UK • P. Vorkas Imperial college London UK • P. Francheshi IASMA Trento Italy
2f7a53630d68c97e5bb0c8cb98f6e2c1.ppt