8a9946861ddfc0bd7c1084c0dbd23d68.ppt
- Количество слайдов: 29

SPIE 2001 (Bi. OS, EI, LASE, Optoelectronics) Report on BIOS conference 4266: Microarrays: Optical Technologies and Informatics S. José (CA), Jan 21 -22 2001 Elisabetta Manduchi

Outline • Organizers – – Michael Bittner (NHGRI) Yidong Chen (NHGRI) Andreas Dorsel (Agilent Technologies) Edward Dougherty (Texas A&M Univ. ) • 1 keynote address, 2 overview papers, 4 half-day sessions: – – Image Data Analysis Detecting Signals: How and What Data Normalization and Quality Control Analysis of Multiple Expression Profiles

Keynote Address David Botstein, Stanford University Gave a general overview on the extraction of biologically useful information from DNA microarray data. Described the work carried out at his and Pat Brown’s lab. Points: • • m. RNA rate of protein synthesis clustering used in their applications: hierarchical and SVD imputation of missing values: nearest neighborhood method cancer studies: use a cocktail of RNAs from different cell lines as the reference (green) channel to compare normal vs tumor complication of a tumor does not mean heterogeneity • statistics is combined with biological knowledge

Overview Papers • Manduchi, Pizarro, Stoeckert (CBIL): RAD as an infrastructure for array data analysis. • Was supposed to be a paper from industry: Basarsky et al. (Axon Instruments): exploring technical limitations for microarray data acquisition and analysis. Substituted by Y. Chen: a general overview similar to (but shorter than) the talk he gave at Penn.
Image Data Analysis • Braendle et al. (Univ. Wien, Novartis Pharma): a generic and robust approach for the analysis of spot array images: – For filter-based arrays but can be applicable to other types. – Grid Fitting • spot amplification (local max values spot centers) • rotation estimation (use projection values) • grid spanning – Quantification • use of the volume of a Gaussian function • correct for overlaps, artifacts, “volcanos”
Image Data Analysis (cont. ) • J. Stein et al. (Nu. Tec Sciences), GLEAMS – – – for radioactivity and 2 -color fluorescence 2 modes: interactive and batch plugs into SLIMS DB uses technologies licensed by: NHGRI, LLNL, IBM main strength: hysteresis methods for target detection and quantification, combining threshold-based segmentation with morphological ideas
Image Data Analysis (cont. ) • Bergemann et al. (Univ. of WA and F. Hutchinson Cancer Research Ctr. ): statistical issues in signal extraction from microarrays – Automated and reliable segmentation (grid alignment and spot detection ellipses of various sizes) – Estimation of background (use mode of histogram, SE computed by bootstrapping) – Flags: if mean signal < upper confidence (point of intersection of bkg curve with signal curve) – Quantification: R|N Binomial (N, p), where N= # hybridized seq. (R and G) p estimated by ave Ri/(Ri+Gi) over the pixels
Image Data Analysis (cont. ) • Hess et al. (Univ. of TX Anderson Cancer Ctr. ): – A general and interesting talk on statistical issues in a microarray core lab, illustrating their approach: • ACCG in-house DB containing: clinical data, microarrays, image, scanning parameters) • Arrays: 4800 spots of which 2304 genes in duplicate, and various controls (positive and blank) • Initial analysis steps: – – – visual inspection plots of intensities and bkg scatterplots (bkg vs intensities, red vs green) bkg subtraction threshold or truncate low values use log transformed values
Image Data Analysis (cont. ) • Hess et al. , ACC (cont. ) • Analysis of replicates: – scatterplots of rep 1 vs rep 2 – plot |diff| vs ave: i. e. |rep 1 -rep 2| vs (rep 1+rep 2)/2 – hexagonal binning, as an aid in visualization • Single-channel replicates (for flagging) – – ignore thresholded points variance decreases with intensity compute a smooth variance function flag points beyond some multiple (76 flags)
Image Data Analysis (cont. ) • Hess et al. , ACC (cont. ) • Analysis of replicate ratios: – as the analysis of single-channel replicates, but use ave of the 4 measurements: (R 1+R 2+G 1+G 2)/4 and plot |log(R 1/G 1)log(R 2/G 2)| against this average – Flag point beyond some multiple (40 flags) • A total of 98 points were flagged (76 from single-channel analysis, 40 from ratio analysis, 18 from both) • Differential expression: – – use ave of the replicate ratios plot ave ratio vs ave intensity flag “significant” ratios “studentized” ratios (need to check on this)
Image Data Analysis (cont. ) • Kegelmeyer et al. (LLNL) described a methodology to calibrate and validate any acquisition and analysis system: – Dilution series to create a gold standard (ground truth). They used 3 positive controls at different dilutions (costant R and G, constant R/varying G, varying R/constant G) – Measure system response characteristics and correct for them (e. g. compensate for cross talk) – Comparative studies of different methods for : preprocessing, bkg subtraction, segmentation, quantitation, normalization
Image Data Analysis (cont. ) • Nadon et al. (Brock Univ. , Canada), very interesting, should follow-up on this (www. imagingresearch. com, also maker of Array. Vision) – – Array. Stat package Novel random error estimation methods Cleaning-up of the data Differential expression: • • pooled error method z-test with correction for multiple testing higher power than standard t-test a more sensitive statistical test (as few as 2 replicates)
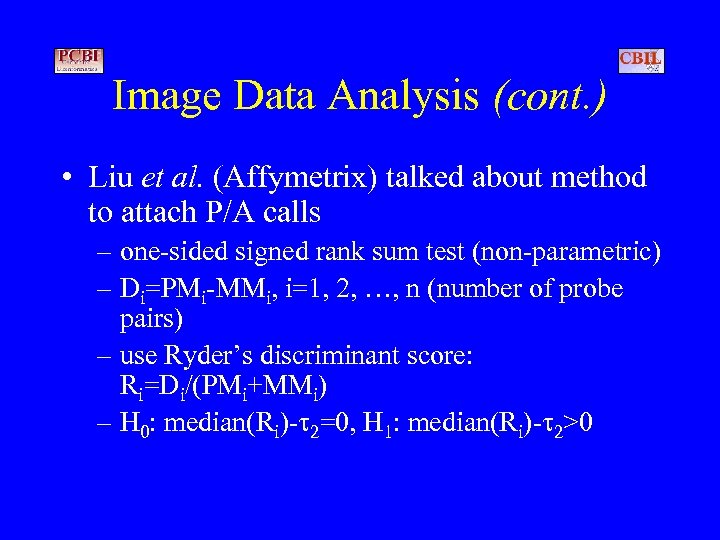
Image Data Analysis (cont. ) • Liu et al. (Affymetrix) talked about method to attach P/A calls – one-sided signed rank sum test (non-parametric) – Di=PMi-MMi, i=1, 2, …, n (number of probe pairs) – use Ryder’s discriminant score: Ri=Di/(PMi+MMi) – H 0: median(Ri)- 2=0, H 1: median(Ri)- 2>0

Detecting Signals: How and What • Guse et al. (Rodenstock Prazisonsoptik, Germany) – Sophisticated lenses for microarray analysis • Shoshan et al. (Compugen) – DNA chips designed to detect alternative splicing • Use a system (LEAD? ) to cluster and assemble ESTs • Build from these suitable probes for oligonucleotide arrays
Detecting Signals (cont. ) Three talks on new labeling methodologies aimed at reducing the amount of RNA needed from the sample (signal amplification) 1. Wong et al. (Quantum Dot and NIH): QD nanocrystals 2. Tyler et al. (NEN Perkins Elmer): tyramide signal amplification (TSA) • Micromax TSA requires 0. 5 -1 g of total RNA vs 50 -100 g required by the direct system

Detecting Signals (cont. ) 3. Getts et al. (Genisphere): dendrimers – – – Hairball (i. e. thumbleweed) structures which can incorporate a lot of fluorescence (250 fluors) Capture sequences for these dendrimers are attached to the c. DNAs Looking into 4 -color analysis Collaborated with V. Cheung People from ACC told me their system did not work for them Another person in the audience commented at the end of the talk that the system was not working for him and he had a hard time to get technical support…
Detecting Signals (cont. ) • Burke et al. (Packard Bio. Chip Tech. ): comparison of labeling methods – both direct and indirect (Cy 3/Cy 5, Amino Allyl, PANVERA’s) – claims the former (which they developed) was cheaper, faster, etc… – Clarified that however they used manufacturer protocols for the others, with no attempt to optimize

Data Normalization and QC • Delenstarr et al. (Agilent Tech. ): very nice talk covering the areas in which Agilent works, with a focus on feature extraction: – – – Probe design c. DNA and oligo microarrays manufacturing Protocol development New generation scanner Feature extraction Bioinformatics (use of Rosetta’s Resolver)
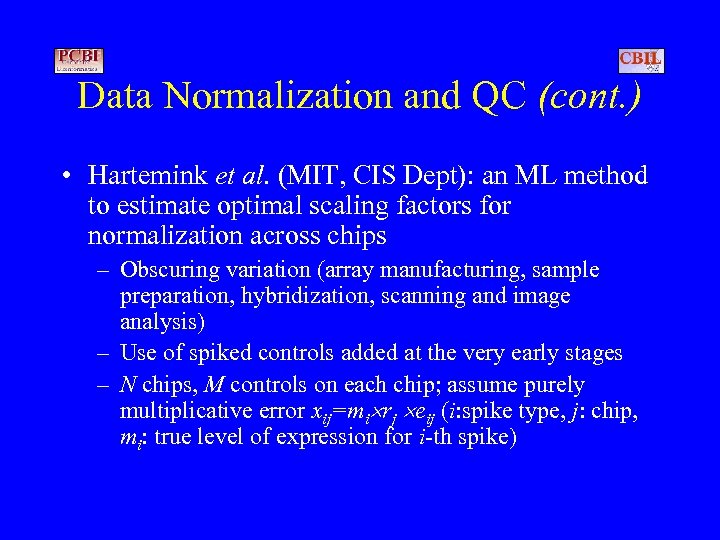
Data Normalization and QC (cont. ) • Hartemink et al. (MIT, CIS Dept): an ML method to estimate optimal scaling factors for normalization across chips – Obscuring variation (array manufacturing, sample preparation, hybridization, scanning and image analysis) – Use of spiked controls added at the very early stages – N chips, M controls on each chip; assume purely multiplicative error xij=mi rj eij (i: spike type, j: chip, mi: true level of expression for i-th spike)
Data Normalization and QC (cont. ) • Hartemink et al. (MIT, CIS Dept) – Log transform yij= i+ j+ ij – Assume ij N(0, i 2), yij N( i+ j, i 2) – Use ML to estimate the 3 parameters of these distribution – Assuming all the spikes of a type have been added in the same amounts, the optimal scaling factor for chip j is a weighted geometric mean of m^i/ xij (the weight depending on the variance) – Future work: incorporate prior over variances, incorporate additive sources of variation
Data Normalization and QC (cont. ) • Yang J. et al. (T. Speed group): a normalization method for c. DNA microarrays where the coefficient depends on the intensities and on the printing tip – Issues: what genes to use, location, scale – Within-slide, paired slides (self-normalization, dye swap), between slides – Assumption: changes roughly symmetric at all intensities
Data Normalization and QC (cont. ) • Luck et al. (Du. Pont Agricultural Genomics): normalization and error estimation for expression patterns • Hsu (National Tsing Hua Univ. , Taiwan): modified confocal scanner system for microarrays
Data Normalization and QC (cont. ) • Coombes (ACC): clustering for quality control – If groups already known, unsupervised clustering can be used for quality control • • data that does not belong poor replicates different filter runs or PCR quality different dynamic ranges – Case study 22, 000 genes, 4 different arrays, 2 replicate experiments per array, several sample types • Preprocessing (bkg subtraction, normalization to median, thresholding, log transformation) • Hierarchical clustering (complete linkage, Euclidean distance) • Experiments clustered where not expected turned out to have quality problems
Data Normalization and QC (cont. ) • Chen et al. (NIH, Texas A&M Univ. ): random signal model for c. DNA microarrays – Simulate microarray images – Simulation tasks • • fluorescent bkg (taken Gaussian) c. DNA target (spot, shape, variable hole) post-processing (scratches, dust, etc. ) image generation (single-channel, multi-channel TIFFs) – Goal: to establish a ground truth for comparison studies and evaluation of packages such methods of image extraction, etc. – Simulation software will be web accessible to the community
Analysis of Multiple Expression Profiles • Alter et al. (Stanford Univ. ): SVD (eigengenes, eigenarrays) • Eilers et al. (Leids Univ. , Netherlands): classification of microarray data with penalized logistic regression – binary classification – logistic regression presents a problem when the number of explanatory variables is large (ML needs #obs>5#var) – used a trick from chemometrics (penalty) and AIC criterion to chose the penalty coefficient.
Analysis of Multiple Expression Profiles • Johnsons et al. (Paradigm Genetics) – Statistical challenges: few replicates, need controls – Can control for systematic variation, but impossible to control for stochastic variation (random error) – Build a model for stochastic error using self-self experiments – Try to understand biological variability for Arabidopsis (standard growth condition, highly reproducible populations, get gene specific error models) – developed a database of baseline characterizations, containing data to serve as support to baseline comparisons
Analysis of Multiple Expression Profiles • 2 talks by Kim et al. (Texas A&M, Univ. de Sao Paulo, NIH) – finding robust linear expression-based classifiers – parallel computing methods for microarray data analysis
Analysis of Multiple Expression Profiles • Dougherty et al. (Texas A&M, Univ. de Sao Paulo, NIH): time series inference from clustering – Need replicates to assess population profile rather than process profile – Studied the effect of # of replicates – Compared clustering methods (k-means, fuzzy c-means, SOM, hierarchical+ Eucl, hierarchical+corr) – First study: 5 synthetic templates from which got simulated replicates (ended with 50 points from each) – Second study: real data, cluster with one method, assume good clusters, seed the model to generate simulated data – Fuzzy c- means performed well (even with 3 replicates), SOM did fine – http: //gspsnap. tamu. edu/clustering (user and pwd=“clustering)”)
Analysis of Multiple Expression Profiles • 2 talks on genetic networks – van Someren et al. (Delft Univ. of Tech. , Netherlands), a comparative study of various models in the 3 categories: pair-wise models, rough networks, complex networks (www. genlab. tudelft. nl) – Barrera et al. (Univ. de Sao Paulo, TX A&M Univ. ): a simulator for gene expression networks
8a9946861ddfc0bd7c1084c0dbd23d68.ppt