b1c3bb9f43877a2dc4f23164139d8b48.ppt
- Количество слайдов: 47

Solving the problem of mixed DNA profiles Dan E. Krane, Wright State University Courtroom Knowledge of Forensic Technology and the Impact on Frye and Daubert Standards Wednesday, August 10, 2016 Forensic Bioinformatics (www. bioforensics. com)
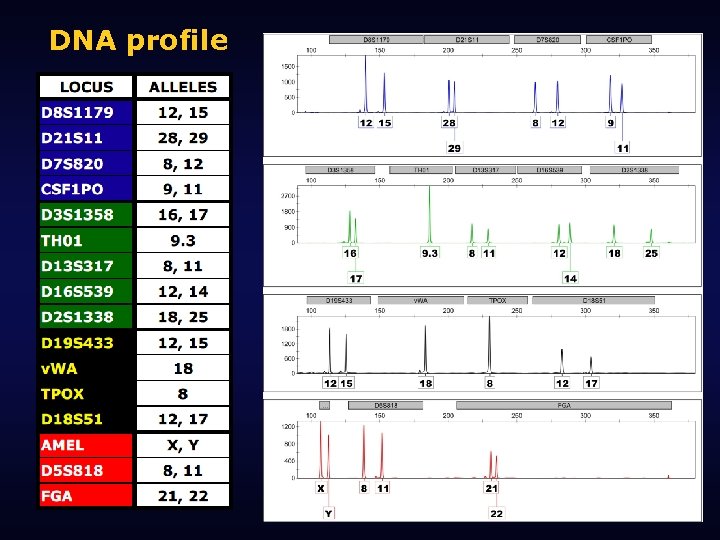
DNA profile
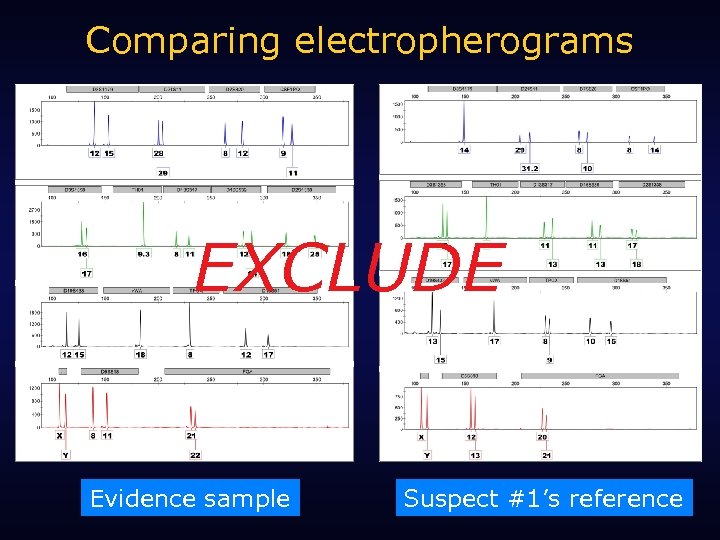
Comparing electropherograms EXCLUDE Evidence sample Suspect #1’s reference
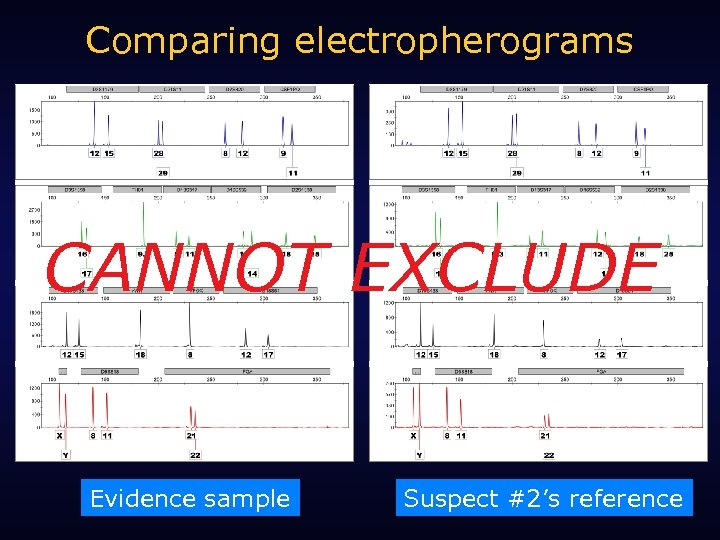
Comparing electropherograms CANNOT EXCLUDE Evidence sample Suspect #2’s reference

What weight should be given to DNA evidence? Statistics do not lie. But, you have to pay close attention to the questions they are addressing. What is the chance that a randomly chosen, unrelated individual from a given population would have the same DNA profile observed in a sample?

Single source statistics: Random Match Probability (RMP)
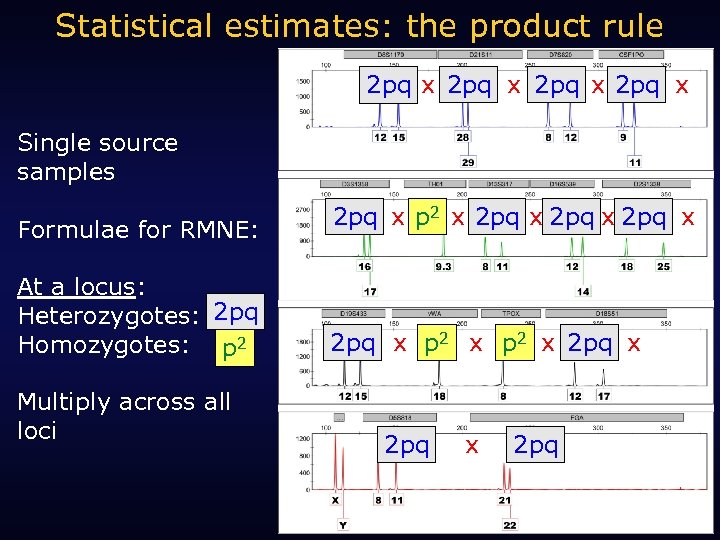
Statistical estimates: the product rule 2 pq x Single source samples Formulae for RMNE: At a locus: Heterozygotes: 2 pq Homozygotes: p 2 Multiply across all loci 2 pq x p 2 x 2 pq x 2 pq
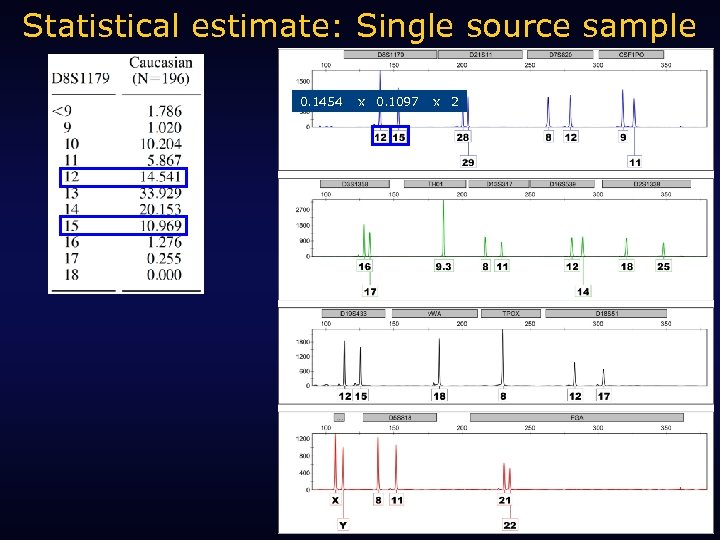
Statistical estimate: Single source sample 0. 1454 x 0. 1097 x 2
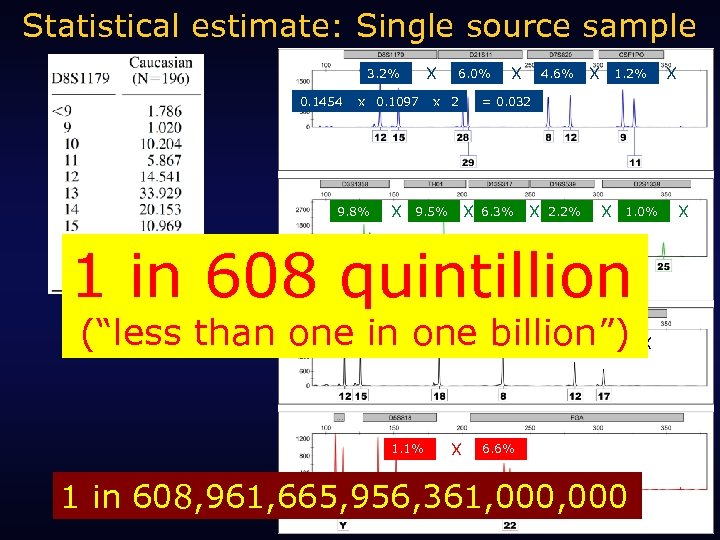
Statistical estimate: Single source sample X 3. 2% 0. 1454 x 0. 1097 9. 8% X 6. 0% x 2 4. 6% X 1. 2% 6. 3% X 2. 2% X 1. 0% 1 in 608 quintillion (“less than one in one billion”) X X 2. 9% 5. 1% 1. 1% X X = 0. 032 X 9. 5% X 29. 9% 4. 0% 6. 6% 1 in 608, 961, 665, 956, 361, 000 X

What weight should be given to DNA evidence? Statistics do not lie. But, you have to pay close attention to the questions they are addressing. What is the chance that a randomly chosen, unrelated individual from a given population would have the same DNA profile observed in a sample?

Mixture statistics: Combined Probability of Inclusion (CPI)
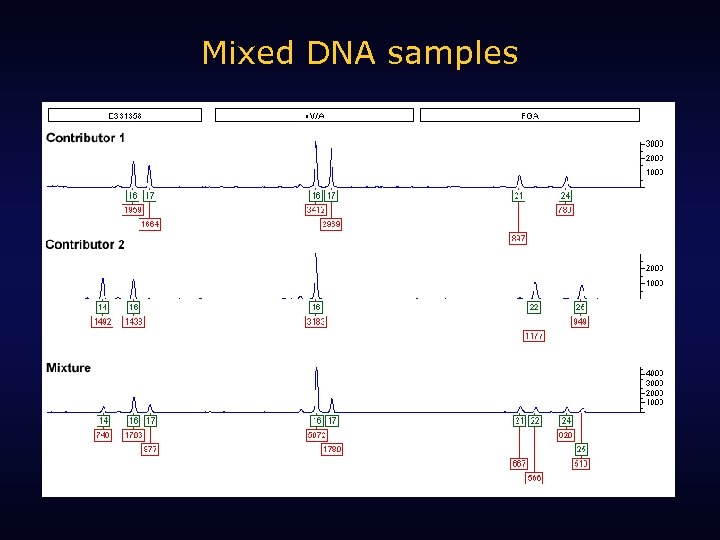
Mixed DNA samples

Put two people’s names into a mixture.

How many names can you take out of this two-person mixture?

How many names can you take out of this two-person mixture?
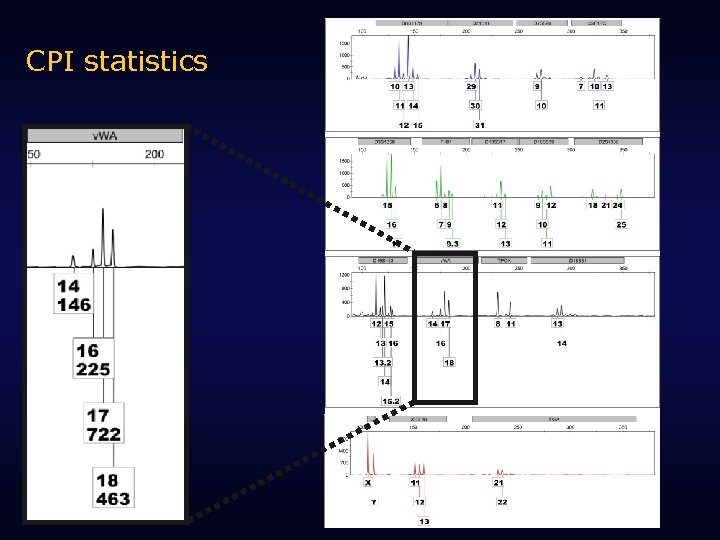
CPI statistics
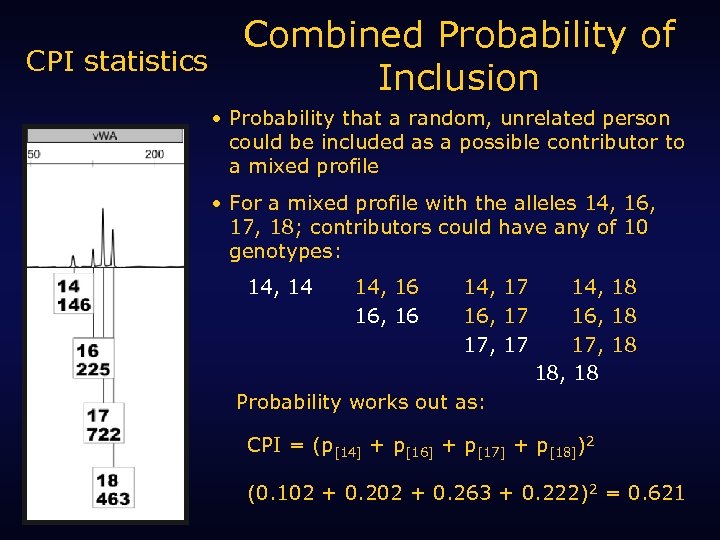
CPI statistics Combined Probability of Inclusion • Probability that a random, unrelated person could be included as a possible contributor to a mixed profile • For a mixed profile with the alleles 14, 16, 17, 18; contributors could have any of 10 genotypes: 14, 14 14, 16 16, 16 14, 17 16, 17 17, 17 14, 18 16, 18 17, 18 18, 18 Probability works out as: CPI = (p[14] + p[16] + p[17] + p[18])2 (0. 102 + 0. 263 + 0. 222)2 = 0. 621
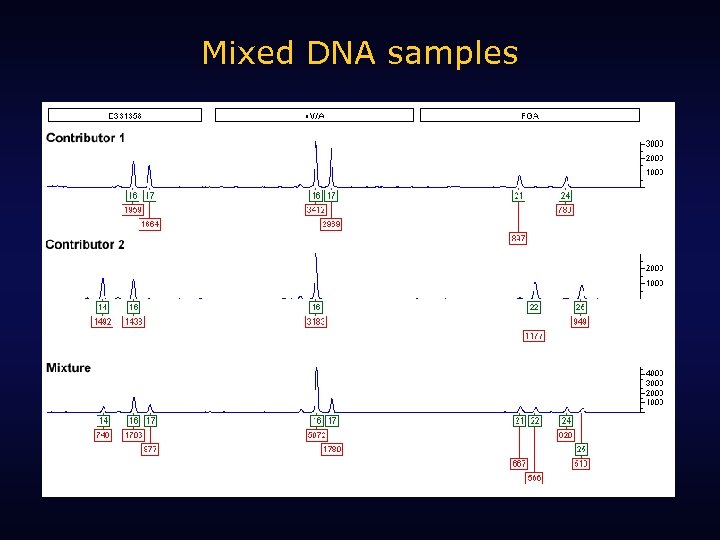
Mixed DNA samples

Mixtures with drop out
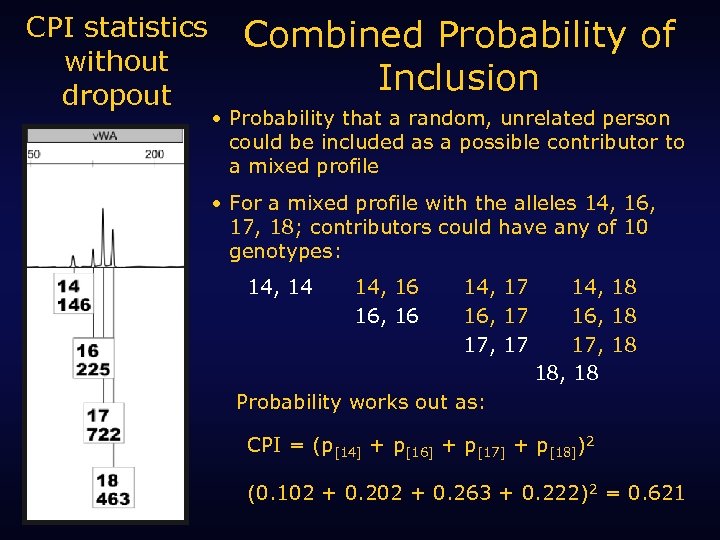
CPI statistics without dropout Combined Probability of Inclusion • Probability that a random, unrelated person could be included as a possible contributor to a mixed profile • For a mixed profile with the alleles 14, 16, 17, 18; contributors could have any of 10 genotypes: 14, 14 14, 16 16, 16 14, 17 16, 17 17, 17 14, 18 16, 18 17, 18 18, 18 Probability works out as: CPI = (p[14] + p[16] + p[17] + p[18])2 (0. 102 + 0. 263 + 0. 222)2 = 0. 621


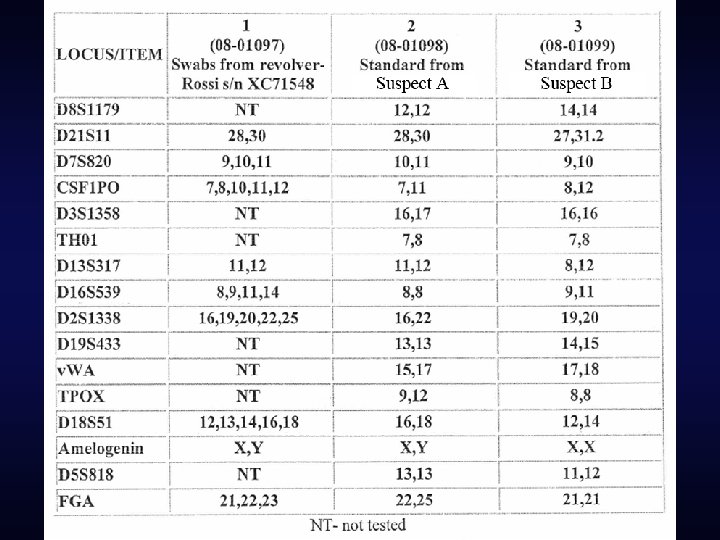

The testing lab’s conclusions
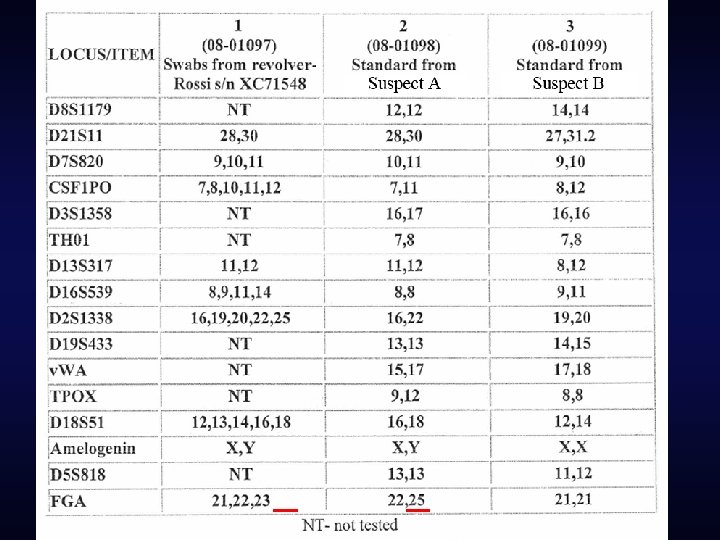
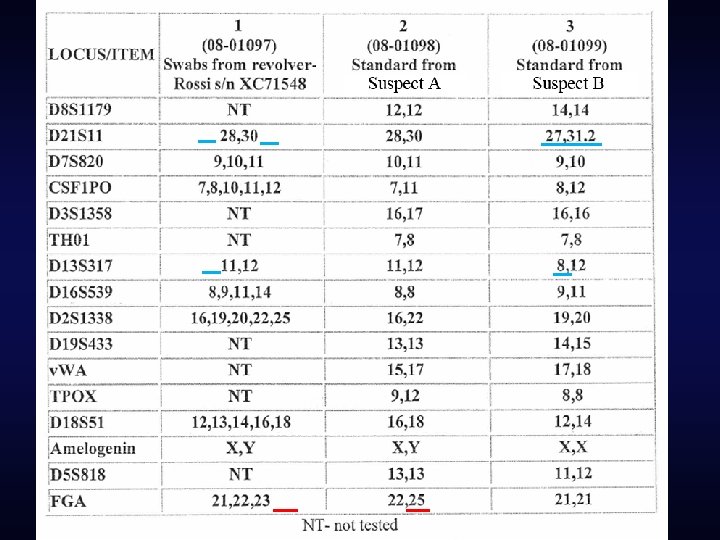

Ignoring loci with “missing” alleles • Some laboratories assert that this is a “conservative” approach • Ignores potentially exculpatory information • “It fails to acknowledge that choosing the omitted loci is suspect-centric and therefore prejudicial against the suspect. ” – Gill, et al. “DNA commission of the International Society of Forensic Genetics: Recommendations on the interpretation of mixtures. ” FSI. 2006.

LCN statistics • No generally accepted method for attaching weight to mixed samples with an unknown number of contributors where dropout may have occurred. • No stats = not admissible.

Why has this become an issue? – More challenging evidence samples • Touch DNA • Guns, steering wheels, doorknobs, etc. – Resulting DNA profiles often: • • Small amounts of DNA Complex mixtures (3 or more persons) Degradation (differential degradation) Minor components in major/minor mixtures – Stochastic effects! – Existing test kits were not designed to test these kinds of samples – Existing statistical methods used in the US are poorly suited to reporting these kinds of samples
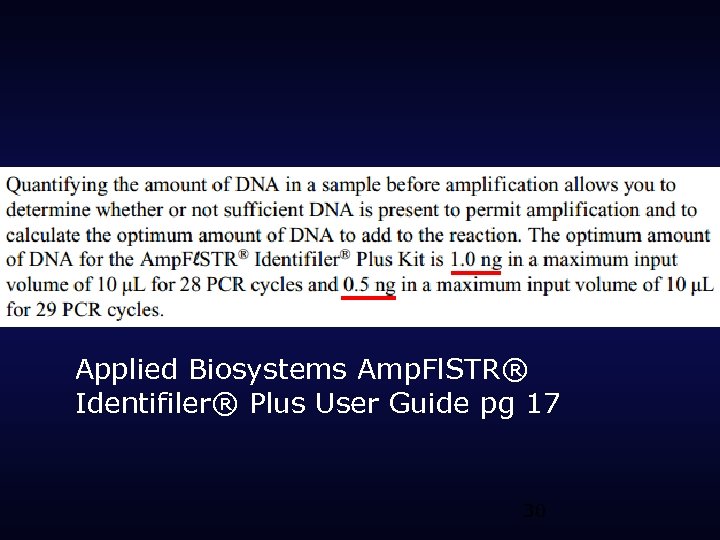
Applied Biosystems Amp. Fl. STR® Identifiler® Plus User Guide pg 17 30

The stochastic threshold • The amount of template DNA where random factors influence test results as much as the actual template. – Exaggerated peak height imbalance – Exaggerated stutter – Allelic drop-in – Allelic drop-out • Sampling error is at the heart of it all
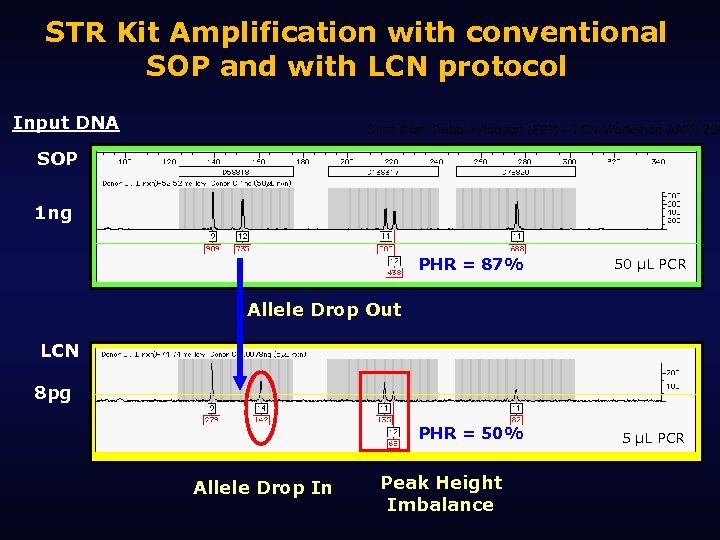
STR Kit Amplification with conventional SOP and with LCN protocol Input DNA Data from Debbie Hobson (FBI) – LCN Workshop AAFS 200 SOP 1 ng PHR = 87% 50 µL PCR PHR = 50% 5 µL PCR Allele Drop Out LCN 8 pg Allele Drop In Peak Height Imbalance
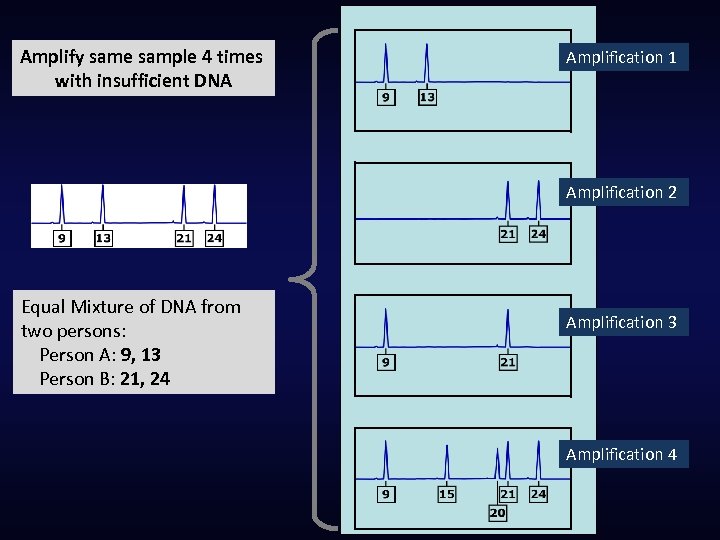
Amplify same sample 4 times with insufficient DNA Amplification 1 Amplification 2 Equal Mixture of DNA from two persons: Person A: 9, 13 Person B: 21, 24 Amplification 3 Amplification 4
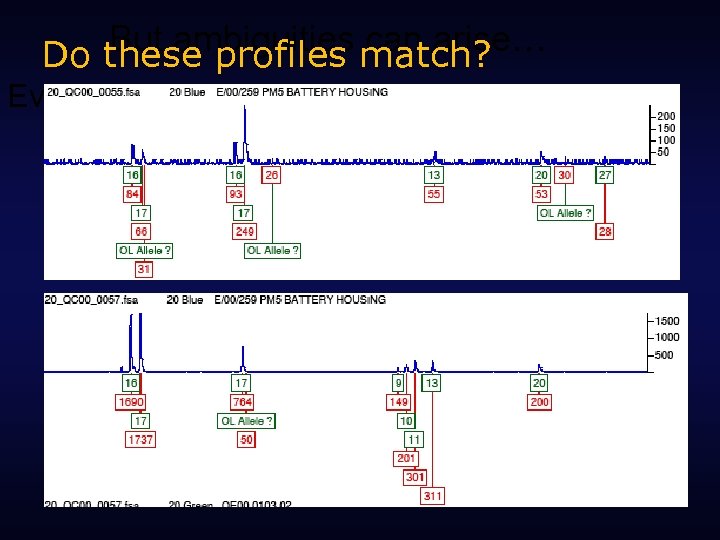
But ambiguities match? can arise… Do these profiles Evidence

Likelihood ratios (LRs) – Compares two alternative hypothesis • “Prosecution” explanation Hp (or H 1) • “Defense” explanation Hd (or H 2) – The likelihood ratio is better able to deal with to continuous data • Enables scientist model stochastic effects and complex mixtures • Complicated – need computer program – Track record: • Widely used in UK, Europe, Australia & New Zealand • Not much in US (other than Paternity Index)
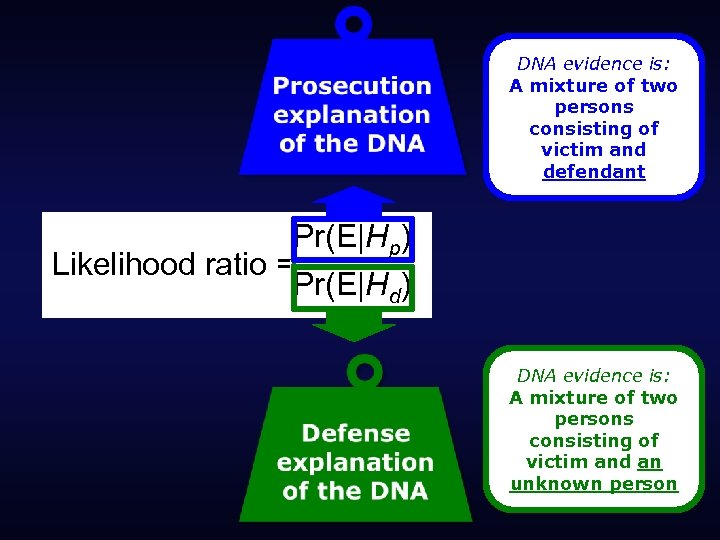
DNA evidence is: A mixture of two persons consisting of victim and defendant Pr(E|Hp) Likelihood ratio = Pr(E|Hd) DNA evidence is: A mixture of two persons consisting of victim and an unknown person
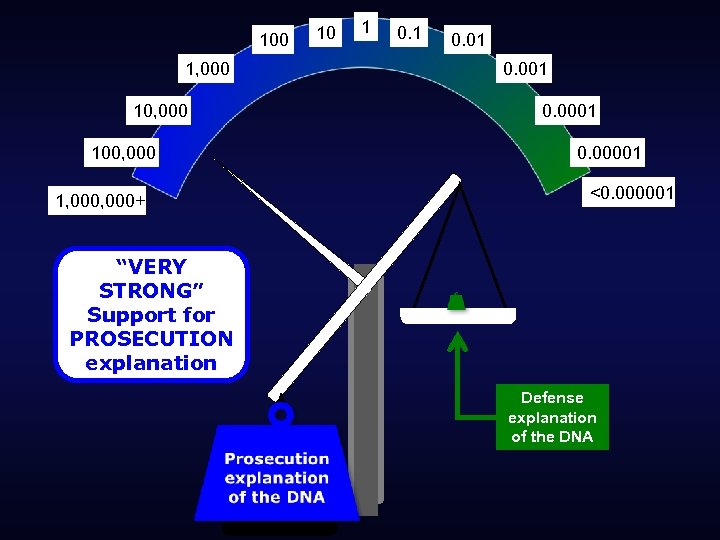
100 1, 000 100, 000 1, 000+ 10 1 0. 01 0. 0001 0. 00001 <0. 000001 “VERY STRONG” Support for PROSECUTION explanation Defense explanation of the DNA

Likelihood Ratio: Drawbacks • Choice of hypotheses can be challenging: – Prosecution Hypothesis (Hp) is usually easy (based on specific allegation) – Defense Hypothesis (Hd) may be more difficult to anticipate • Can do multiple pairs of hypotheses • In mixtures need to specify number of contributors – Can have different numbers of contributors in Hp and Hd • Always look at the hypotheses carefully to check they accurately represent the facts of the case

Why do we need probabilistic genotyping? – More challenging evidence samples • Touch DNA • Guns, steering wheels, doorknobs, etc. – Resulting DNA profiles often: • • Small amounts of DNA Complex mixtures (3 or more persons) Degradation (differential degradation) Minor components in major/minor mixtures – Stochastic effects! Existing statistical methods used in the US are poorly suited to reporting these kinds of samples
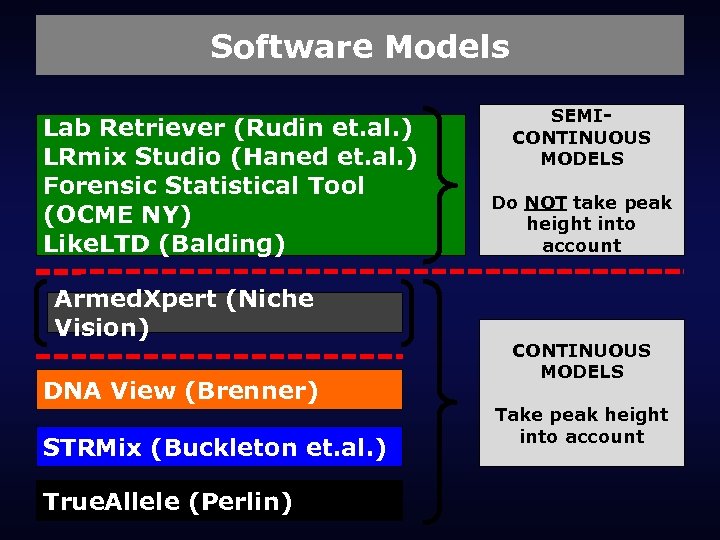
Software Models Lab Retriever (Rudin et. al. ) LRmix Studio (Haned et. al. ) Forensic Statistical Tool (OCME NY) Like. LTD (Balding) Armed. Xpert (Niche Vision) DNA View (Brenner) STRMix (Buckleton et. al. ) True. Allele (Perlin) SEMICONTINUOUS MODELS Do NOT take peak height into account CONTINUOUS MODELS Take peak height into account

So, what do most of these programs do (… in plain language)? Part I • • Run DNA test (as usual) – resulting in e-data Analyze electronic data with Gene. Mapper ID (as usual) Review electropherograms (as usual) Interpret (as usual) – Decide on MATCHES, EXCLUSIONS and INCONCLUSIVES USUALLY AT THIS STAGE ANALYST WOULD USE POPSTATS TO CALCULATE STATS AND THEN WRITE REPORT • Consider the LR hypotheses you may want to use – Victim present? – Number of contributors? • Return to Gene. Mapper and prepare a special tabular export of the allele calls (including peak heights) for the evidence sample and refs. that you want to compare – Remove artifacts and rare alleles – May or may not include stutter peaks – May drop analytical threshold to a lower level to capture more peaks

So, what do most of these programs do (… in plain language)? Part II • Import tabular data into Probabilistic Software • Frame LR Hypotheses, for example: – HP = VICTIM plus DEFENDANT plus ONE UNKNOWN PERSON – Hd = VICTIM plus TWO UNKNOWN PERSONS • Set drop-out estimate – Methods differ in how this is done – May be based on the data – May be flat estimate • Set drop-in estimate – Usually use flat estimate • Set up additional variables – Depends of software program • Run program! • Review output • Program will give a numerical value indicating the Likelihood Ratio – If above 1, supports prosecution hypothesis – If below 1, supports defense hypothesis – Inconclusive range around 1

This is true for most of the programs, but True. Allele is different • • Run DNA test (as usual) – resulting in e-data Analyze electronic data with Gene. Mapper ID (as usual) Review electropherograms (as usual) Interpret (as usual) True. Allele USUALLY AT THIS STAGE ANALYST WOULD USE POPSTATS TO DOES THE REST CALCULATE STATS AND THEN WRITE REPORT (and the LR hypotheses you may want to use page most of the other • Consider – as well) – – Decide on MATCHES, EXCLUSIONS and INCONCLUSIVES Victim present? Number of contributors? • Return to Gene. Mapper and prepare a special tabular export of the allele calls (including peak heights) for the evidence sample and refs. that you want to compare – Remove artifacts and rare alleles – May or may not include stutter peaks – May drop analytical threshold to a lower level to capture more peaks

True. Allele – Continuous approach • Models peak heights • Uses MCMC – Imports raw electronic data – Uses its own smoothing (not Gene. Mapper) • Perlin says it is “equivalent” to ABI’s data in terms of peak heights • But peak heights are not the same – True. Allele performs all the analysis of the data • Including the Gene. Mapper analysis usually done by the lab analyst – True. Allele is intended to replace the analyst • Interpret the data • Make the “matches” • Calculate the statistics
True. Allele – Models 100 s of variables: • Some are known, such as degradation and relative amounts of DNA: • The vast majority have not been described – Uses a very low analytical threshold (10 RFU) – Unlike STRMix and other approaches, True. Allele does not need a lab or test kit-specific variance factor – The program is able to take into account such things as: • • • Stutter (plus and minus) Biochemical and electrical artifacts Type of test (Identifiler, Profiler etc. ) Type of instrument (3130, 3500) What else?

True. Allele – Proponents say that validation studies show that it ”gets the right answers”: • Known mixtures rarely have LRs for known non-contributors that are greater than those for known contributors • Several peer-reviewed papers outline general approach – Detractors worry about the black box and failure to define limitations: • At least a dozen hotly debated questions must have been resolved to generate a reliable result • Software engineering concerns/right to confrontation • Validation studies do find known non-contributors with positive LRs • No clear features of samples for which True. Allele is known to not generate reliable results

Solving the problem of mixed DNA profiles Dan E. Krane, Wright State University Courtroom Knowledge of Forensic Technology and the Impact on Frye and Daubert Standards Wednesday, August 10, 2016 Forensic Bioinformatics (www. bioforensics. com)
b1c3bb9f43877a2dc4f23164139d8b48.ppt