75361a5d9867b9bba7ac8b46256053fb.ppt
- Количество слайдов: 54
Software and Databases for managing and selecting molecular markers General introduction Pathway approach for candidate gene identification and introduction to metabolic pathway databases. Identification of polymorphisms in data-based sequences

Databases (General and Crop Specific) Germplasm GRIN: http: //www. ars-grin. gov/npgs/ TGRC: http: //tgrc. ucdavis. edu/ Sequence NCBI: http: //www. ncbi. nlm. nih. gov/ SGN: http: //solgenomics. net/ Metabolic Plant. Cyc: http: //www. plantcyc. org: 1555/PLANT/server. html?
New format to NCBI
Access current and past scientific lit.
Increased emphasis on phenotypic data
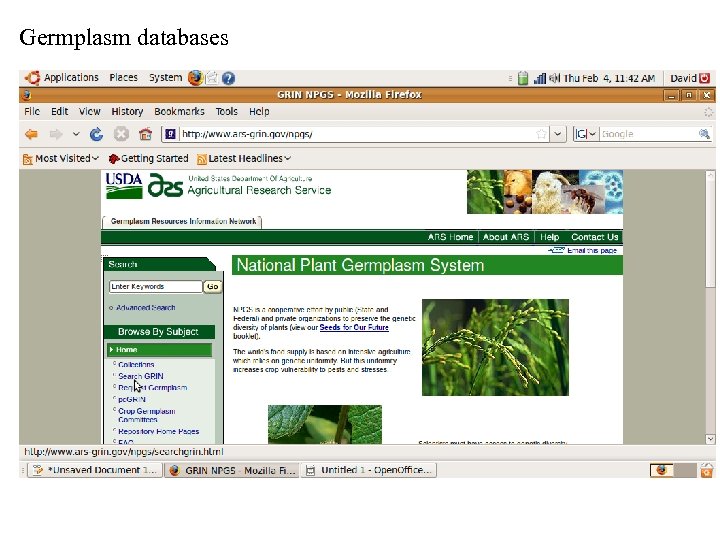
Germplasm databases
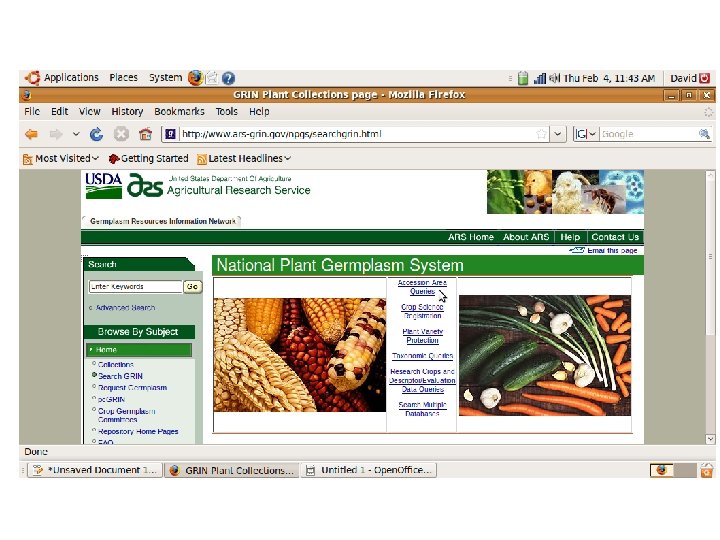
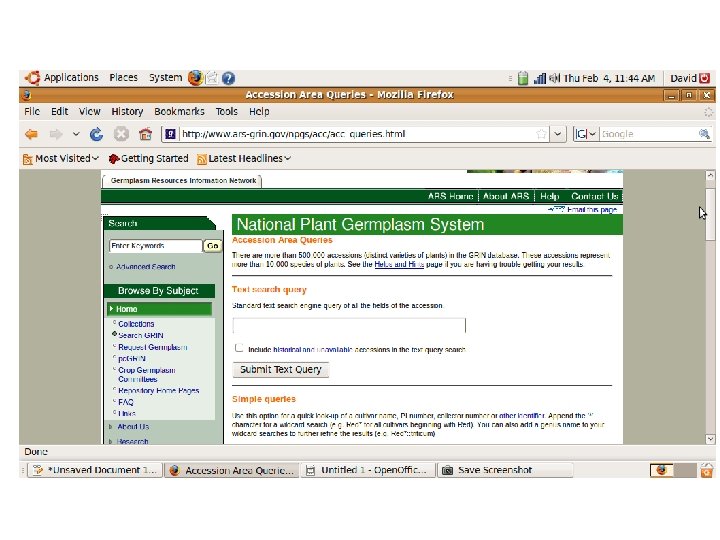
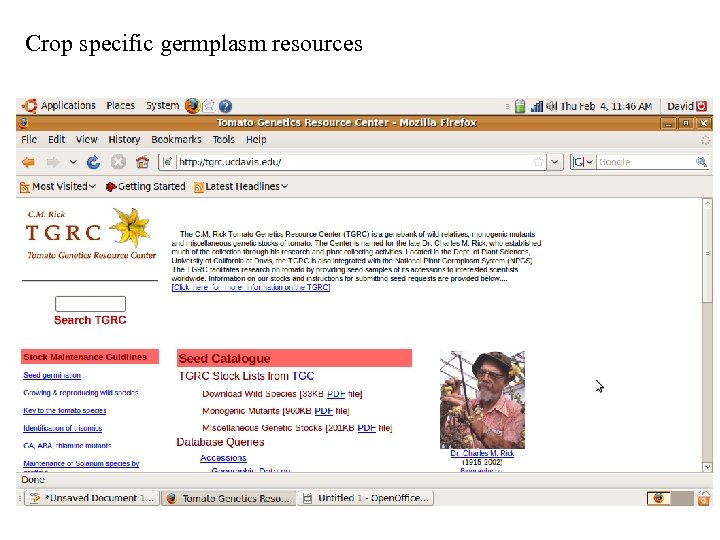
Crop specific germplasm resources
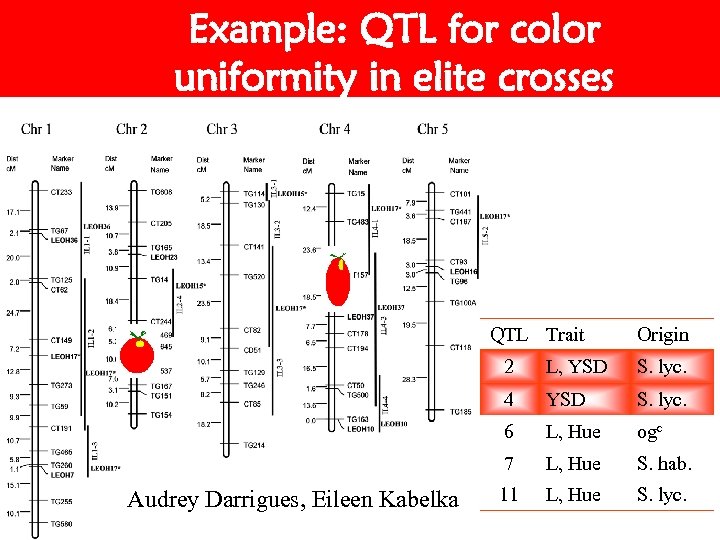
Example: QTL for color uniformity in elite crosses QTL Trait Origin 2 S. lyc. 4 YSD S. lyc. 6 L, Hue ogc 7 Audrey Darrigues, Eileen Kabelka L, YSD L, Hue S. hab. 11 L, Hue S. lyc.
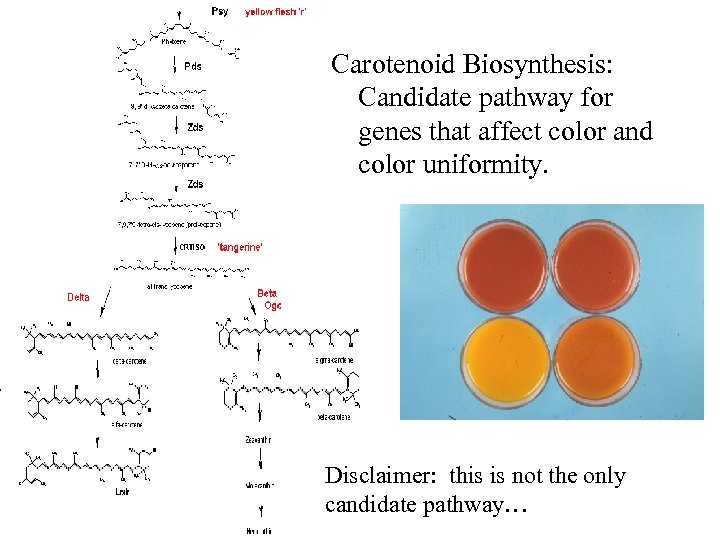
Carotenoid Biosynthesis: Candidate pathway for genes that affect color and color uniformity. Disclaimer: this is not the only candidate pathway…
Databases that link pathways to genes http: //www. arabidopsis. org/help/tutorials/aracyc_intro. jsp

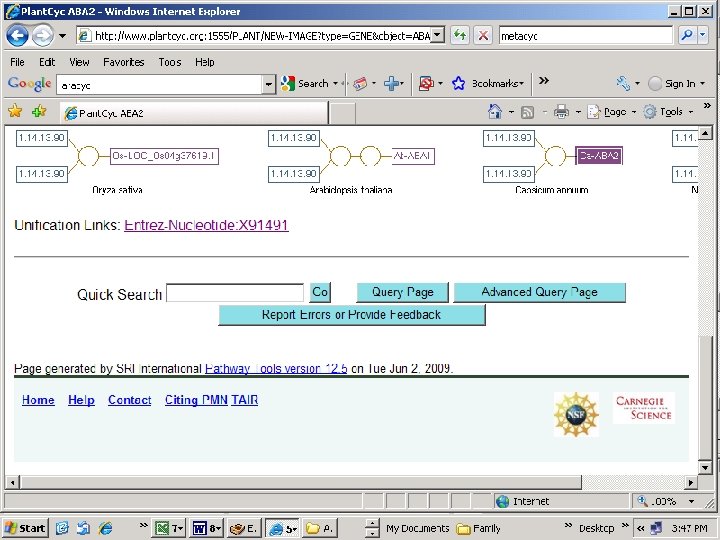
Databases that link pathways to genes http: //metacyc. org/ http: //www. plantcyc. org/ http: //sgn. cornell. edu/tools/solcyc/ http: //www. arabidopsis. org/biocyc/index. jsp http: //www. arabidopsis. org/help/tutorials/aracyc_intro. jsp External Plant Metabolic databases Cap. Cyc (Pepper) (C. anuum) Coffea. Cyc (Coffee) (C. canephora) Sol. Cyc (Tomato) (S. lycopersicum) Nicotiana. Cyc (Tobacco) (N. tabacum) Petunia. Cyc (Petunia) (P. hybrida) Potato. Cyc (Potato) (S. tuberosum) Sola. Cyc (Eggplant) (S. melongena)
http: //www. plantcyc. org: 1555/
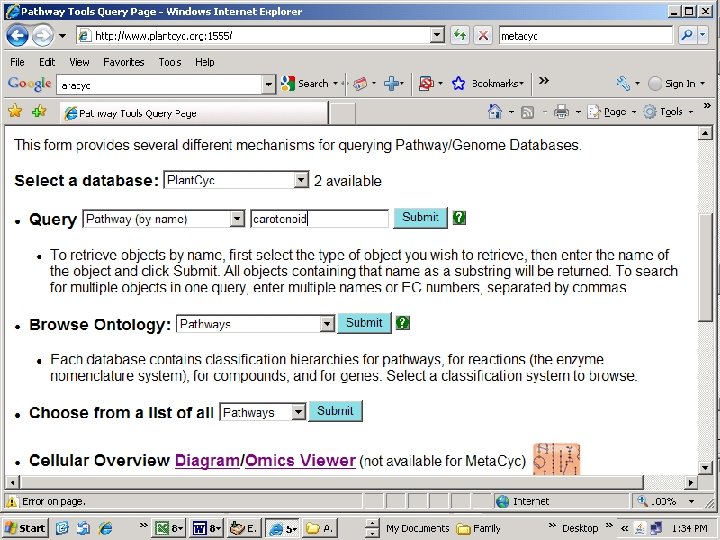
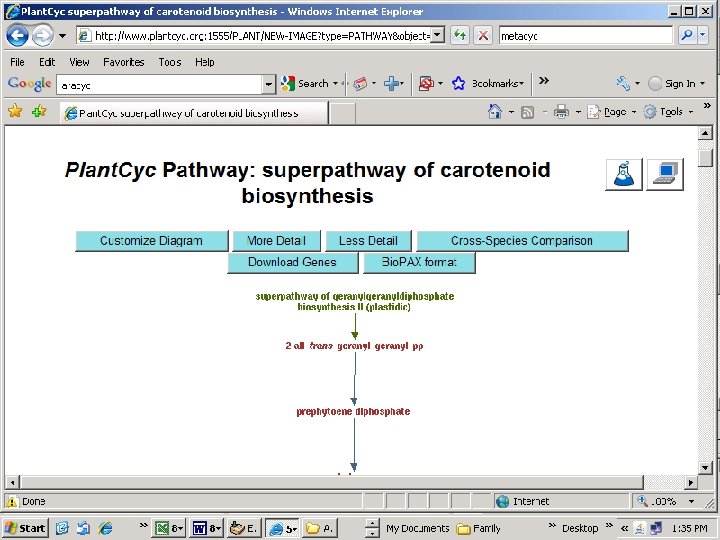
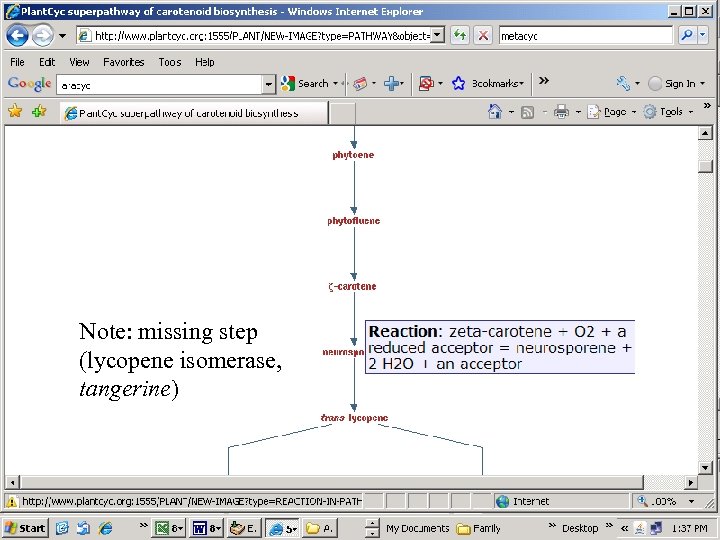
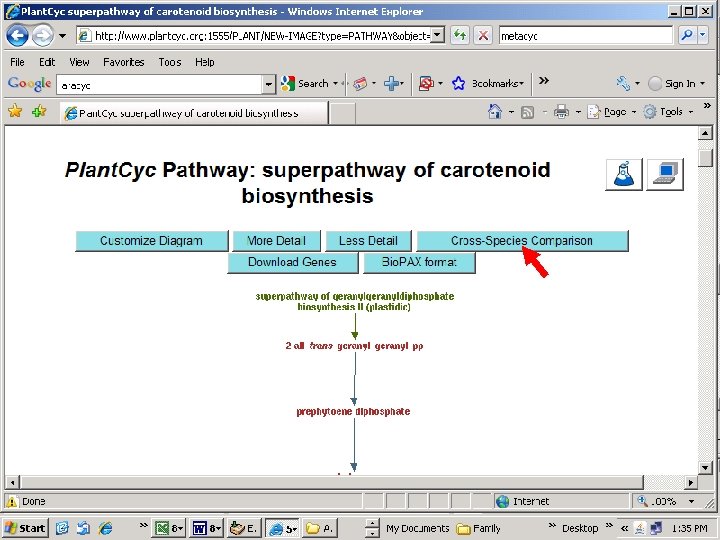
Note: missing step (lycopene isomerase, tangerine)
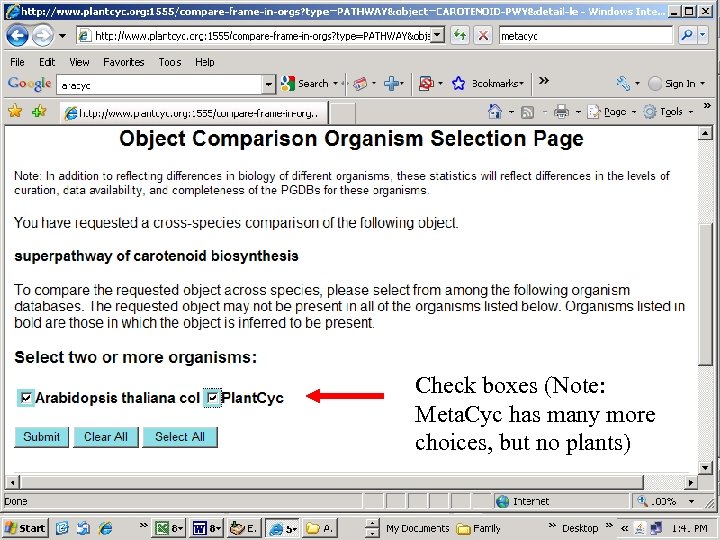
Check boxes (Note: Meta. Cyc has many more choices, but no plants)
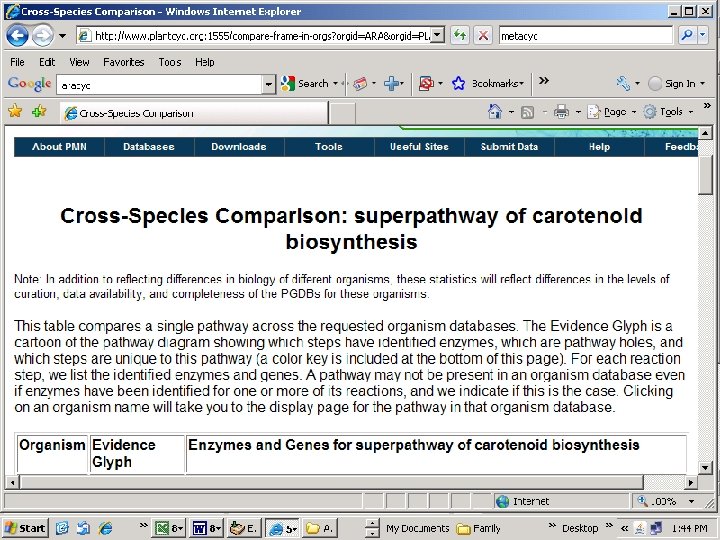
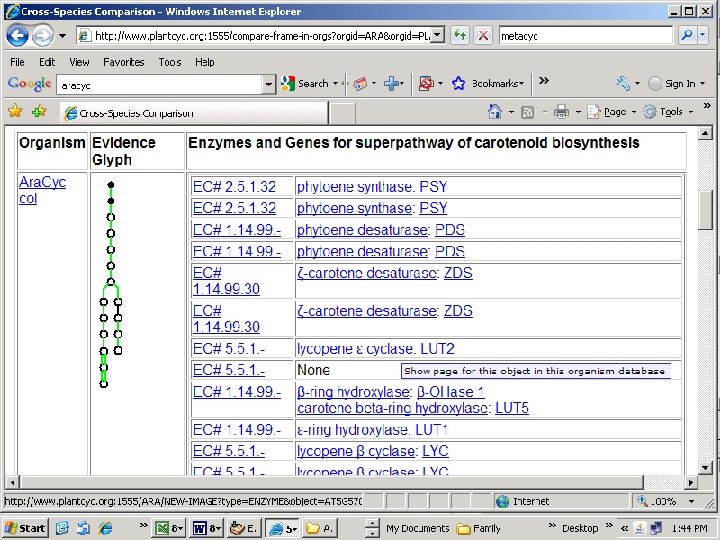
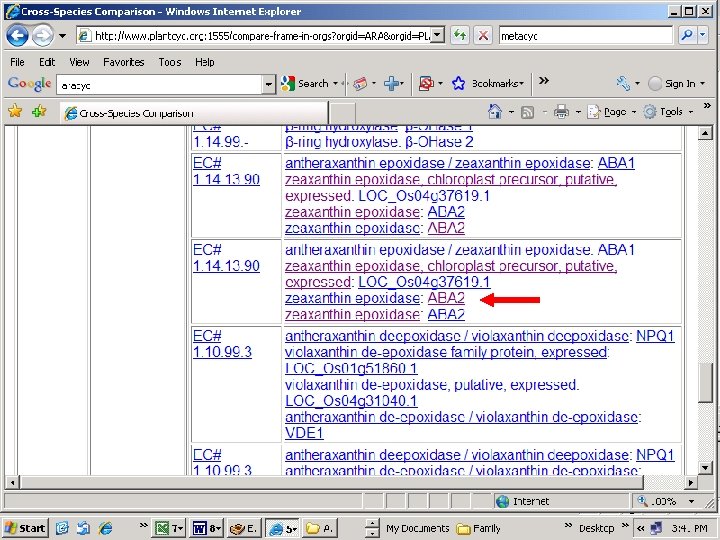
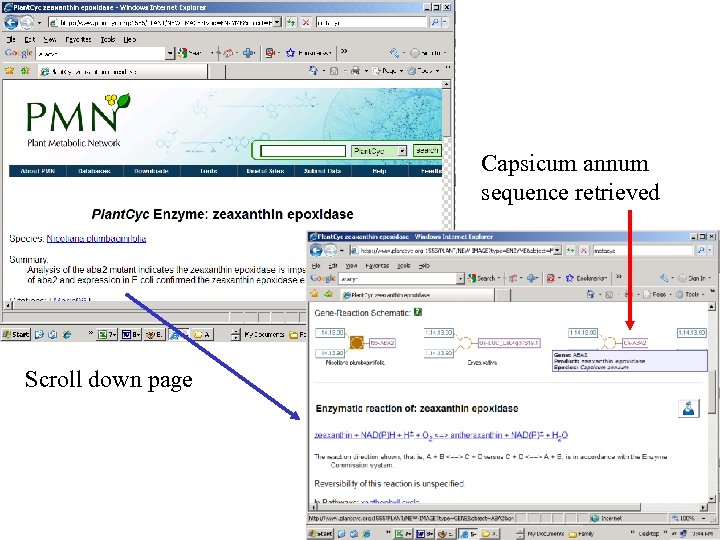
Capsicum annum sequence retrieved Scroll down page
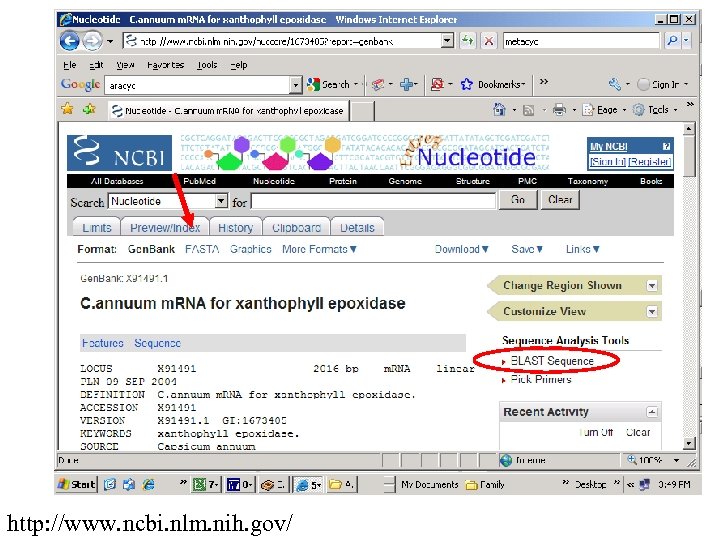
http: //www. ncbi. nlm. nih. gov/
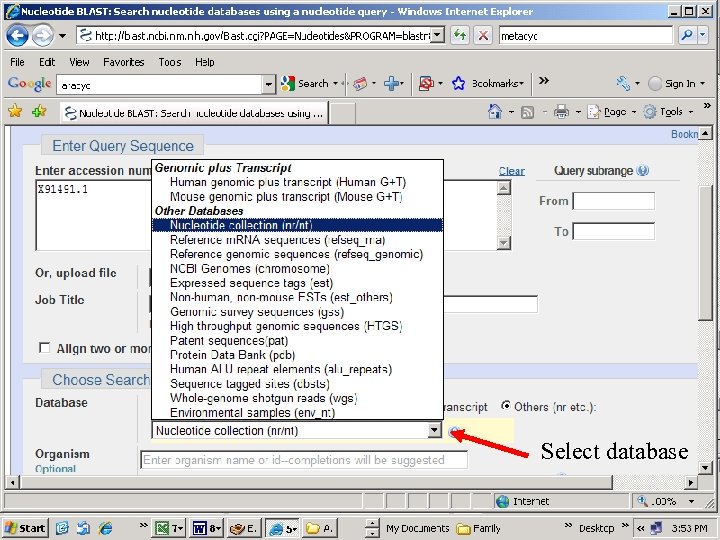
Select database
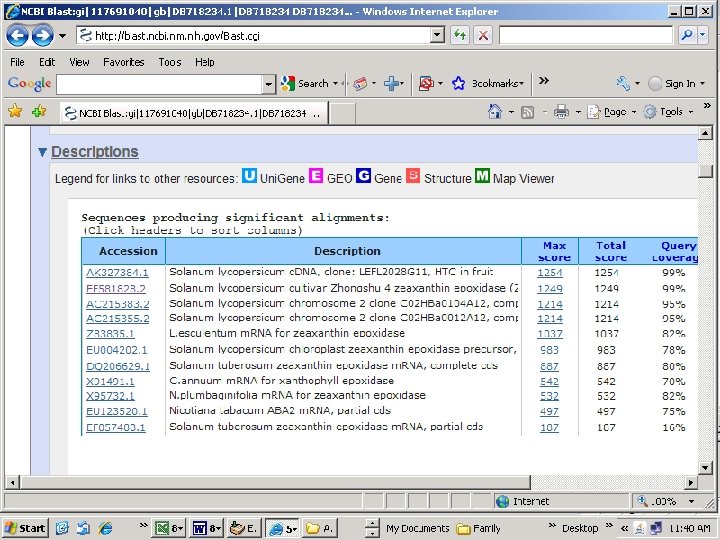

Query CCACCACCATCCTCACTTTAACCCACAAATCCCACTTTGGCCTAATTAACAATTTT ||||||||||||||||| Sbjct CCACCACCATCCTCACTTTAACCCACAAATCCCATTTTCTTTGGCCTAATTAACAATTTT Zeaxanthin epoxidase Probable location on Chromosome 2 Alignment of Z 83835 and EF 581828 reveals 5 SNPs over ~2000 bp
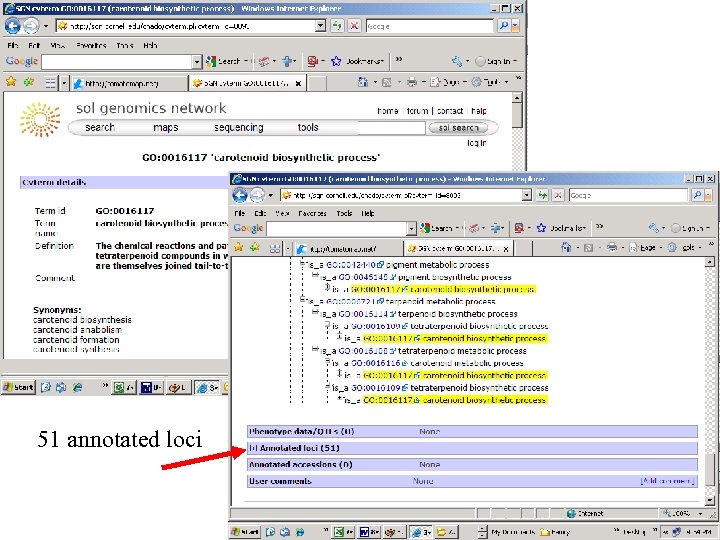
51 annotated loci
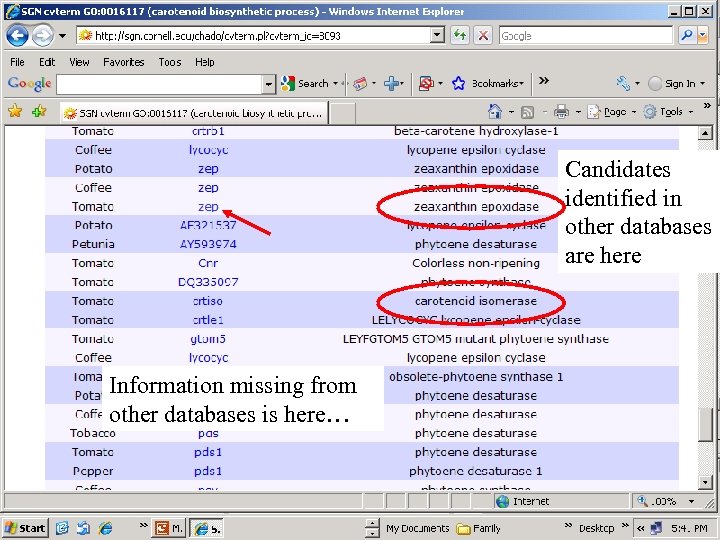
Candidates identified in other databases are here Information missing from other databases is here…
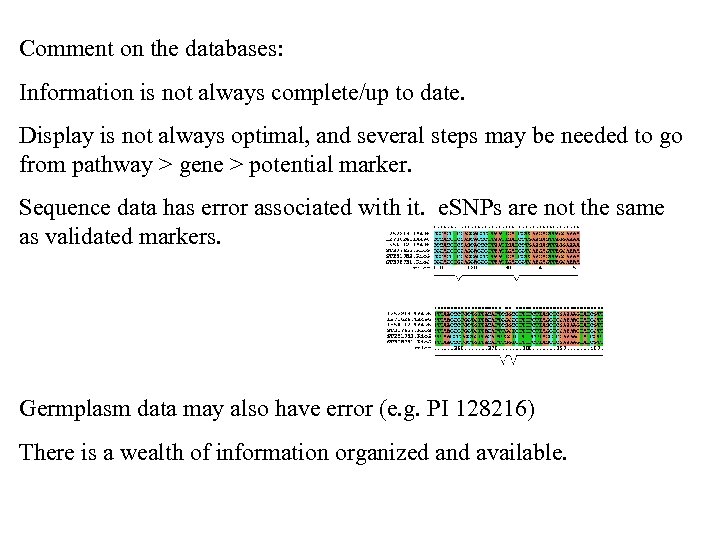
Comment on the databases: Information is not always complete/up to date. Display is not always optimal, and several steps may be needed to go from pathway > gene > potential marker. Sequence data has error associated with it. e. SNPs are not the same as validated markers. Germplasm data may also have error (e. g. PI 128216) There is a wealth of information organized and available.

The previous example detailed how we might identify sequence based markers for trait selection. Query CCACCACCATCCTCACTTTAACCCACAAATCCCACTTTGGCCTAATTAACAATTTT ||||||||||||||||| Sbjct CCACCACCATCCTCACTTTAACCCACAAATCCCATTTTCTTTGGCCTAATTAACAATTTT Improving efficiency of selection in terms of 1) relative efficiency of selection, 2) time, 3) gain under selection and 4) cost will benefit from markers for both forward and background selection. Remainder of Presentation will focus on Where to apply markers in a program Forward and background selection Marker resources Alternative population structures and size
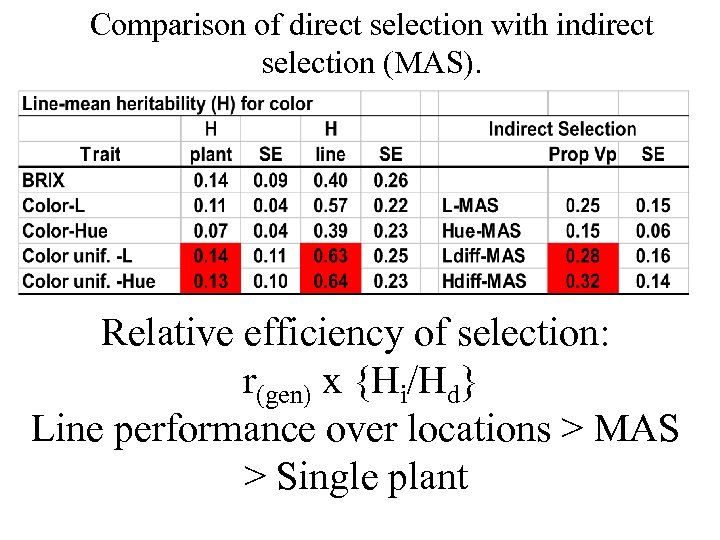
Comparison of direct selection with indirect selection (MAS). Relative efficiency of selection: r(gen) x {Hi/Hd} Line performance over locations > MAS > Single plant
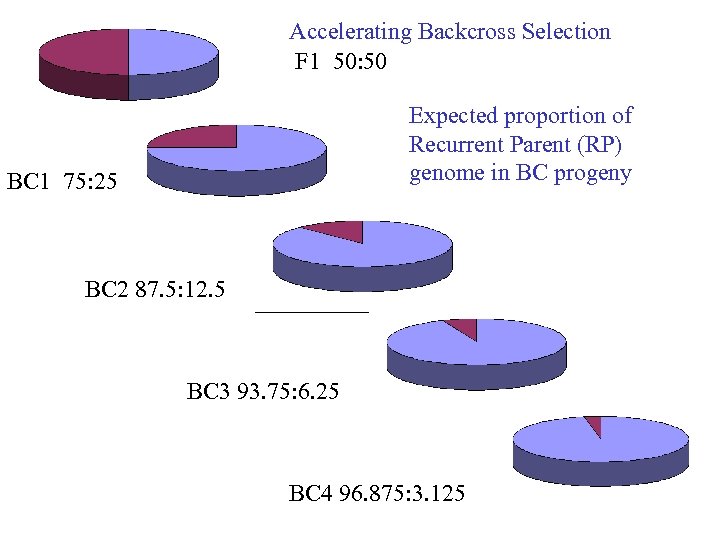
Accelerating Backcross Selection F 1 50: 50 Expected proportion of Recurrent Parent (RP) genome in BC progeny BC 1 75: 25 BC 2 87. 5: 12. 5 BC 3 93. 75: 6. 25 BC 4 96. 875: 3. 125
References: Frisch, M. Bohn, and A. E. Melchinger. 1999. Comparison of Selection Strategies for Marker-Assisted Backcrossing of a Gene. Crop Science 39: 1295 -1301.
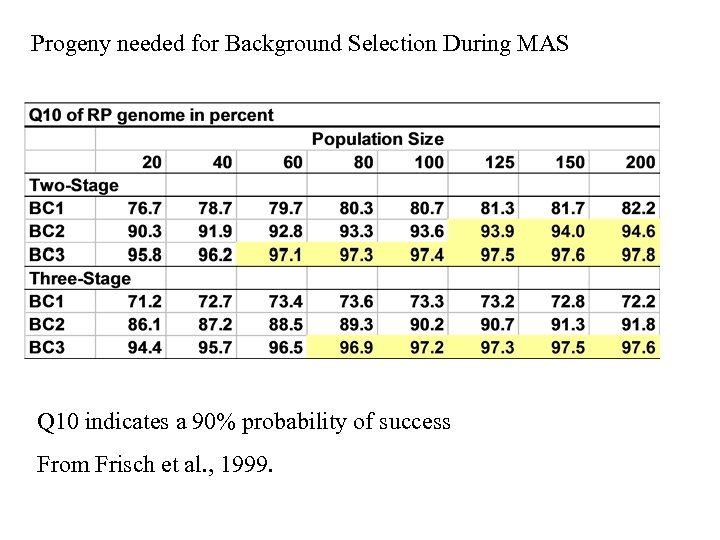
Progeny needed for Background Selection During MAS Q 10 indicates a 90% probability of success From Frisch et al. , 1999.
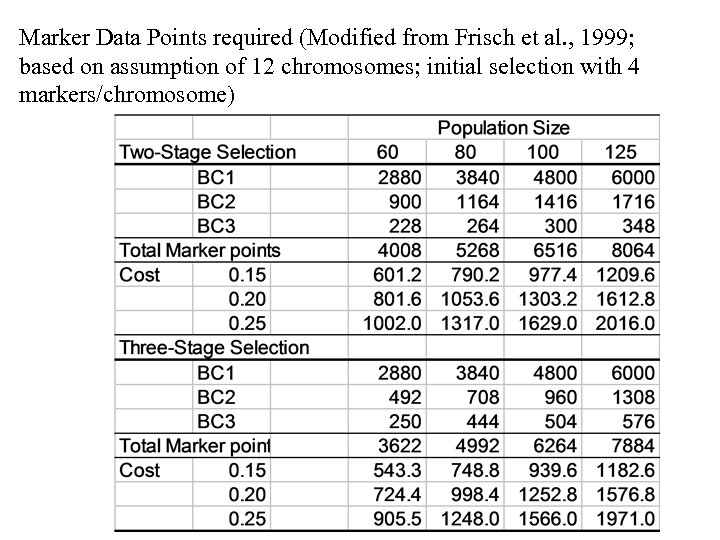
Marker Data Points required (Modified from Frisch et al. , 1999; based on assumption of 12 chromosomes; initial selection with 4 markers/chromosome)

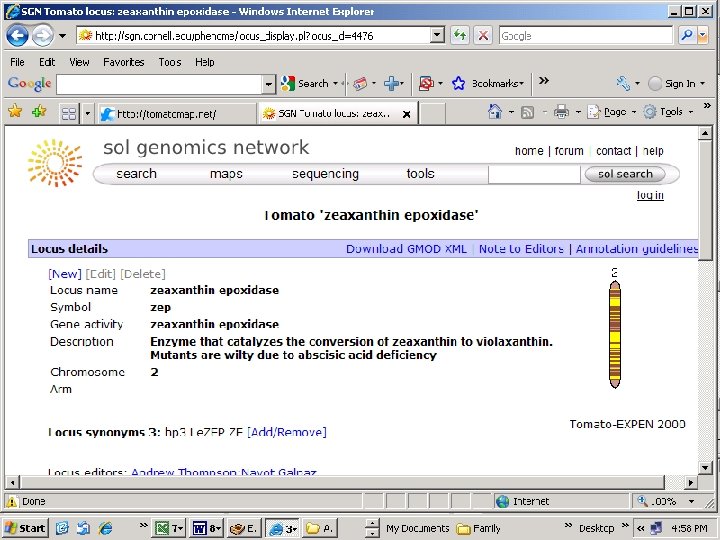
For effective background selection we need: Markers for our target locus (C > T SNP for Zep) Markers on the target chromosome (Chrom. 2) Markers unlinked to the target chromosome (~2 per chromosome arm)
http: //www. tomap. net http: //sgn. cornell. edu/
Ovate
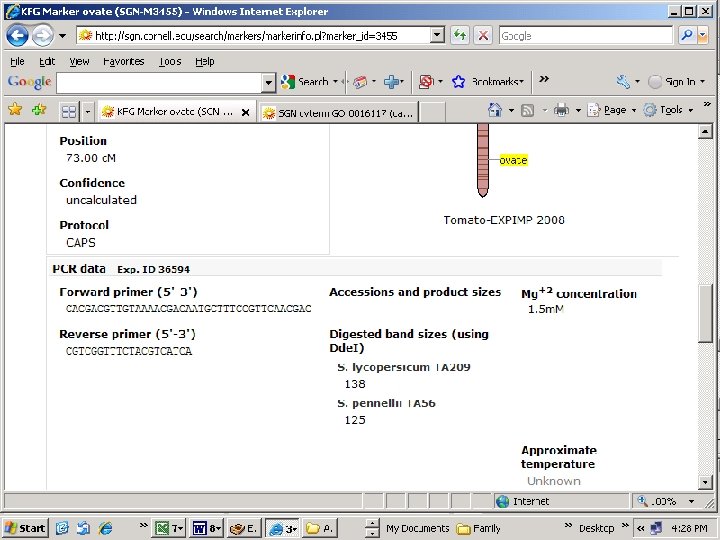
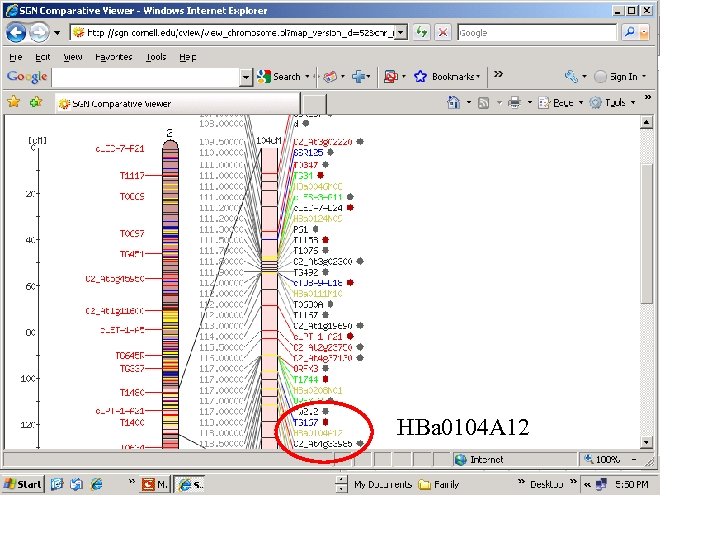
HBa 0104 A 12
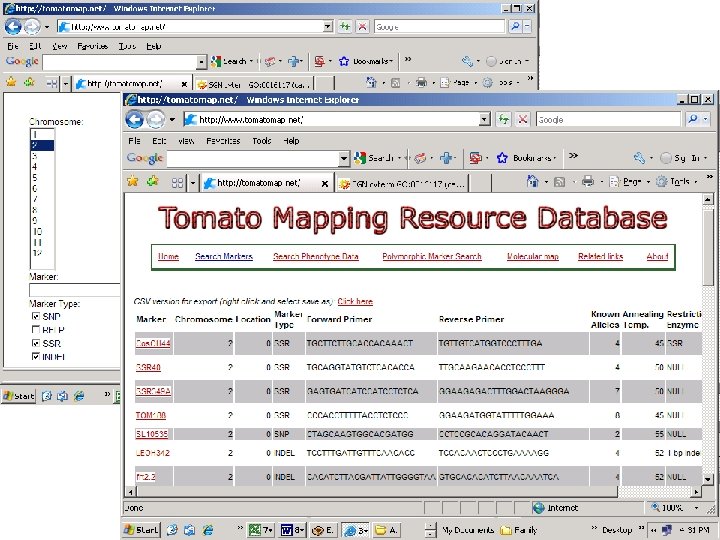
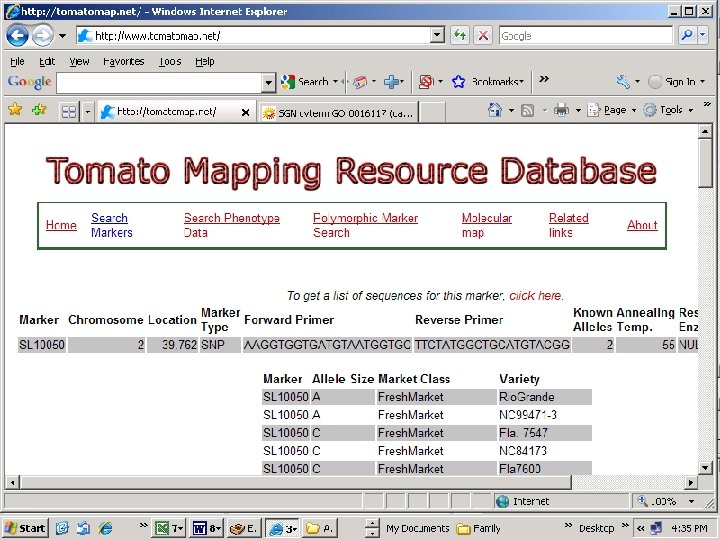
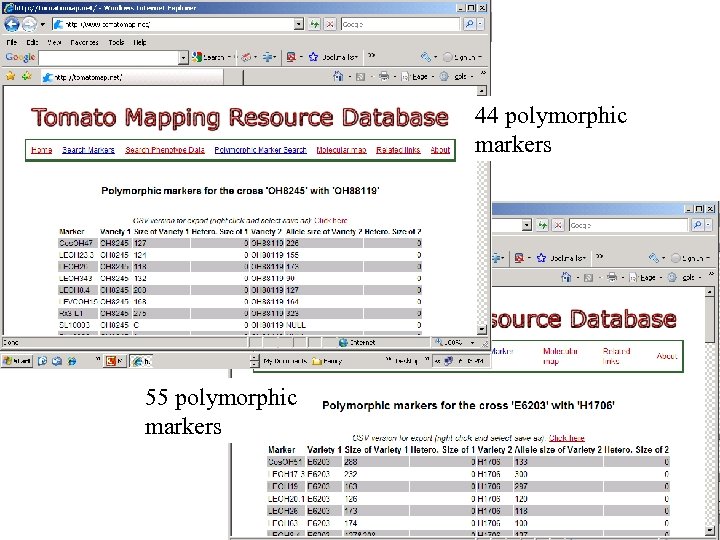
44 polymorphic markers 55 polymorphic markers
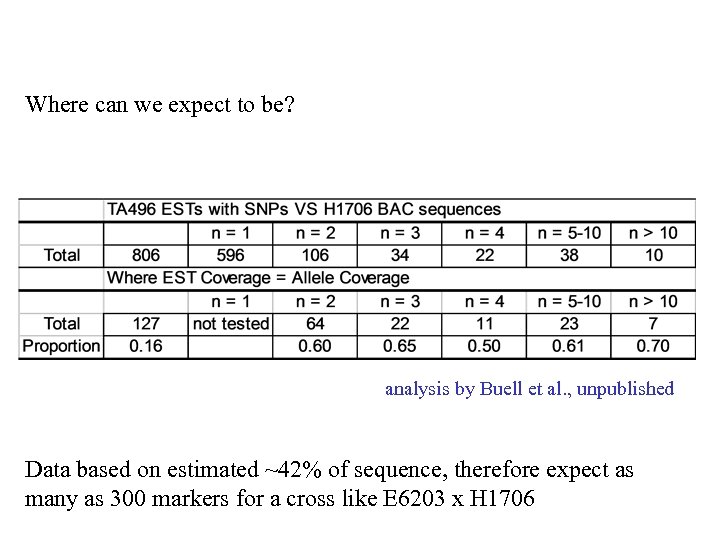
Where can we expect to be? analysis by Buell et al. , unpublished Data based on estimated ~42% of sequence, therefore expect as many as 300 markers for a cross like E 6203 x H 1706
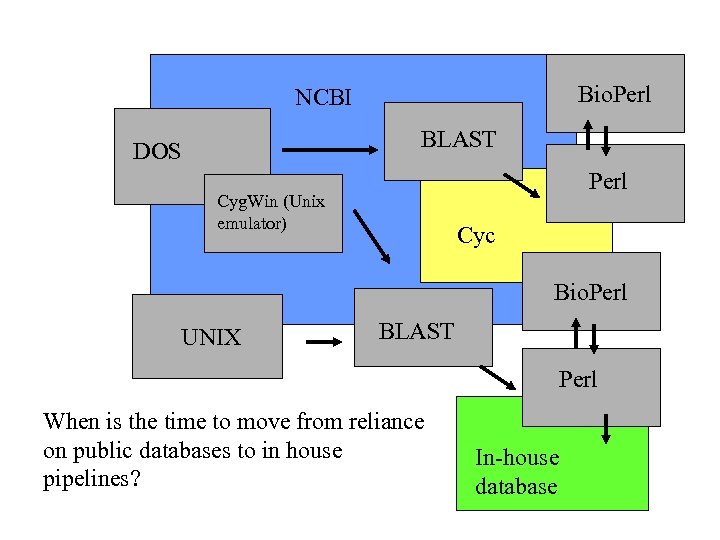
Bio. Perl NCBI BLAST DOS Perl Cyg. Win (Unix emulator) Cyc Bio. Perl UNIX BLAST Perl When is the time to move from reliance on public databases to in house pipelines? In-house database
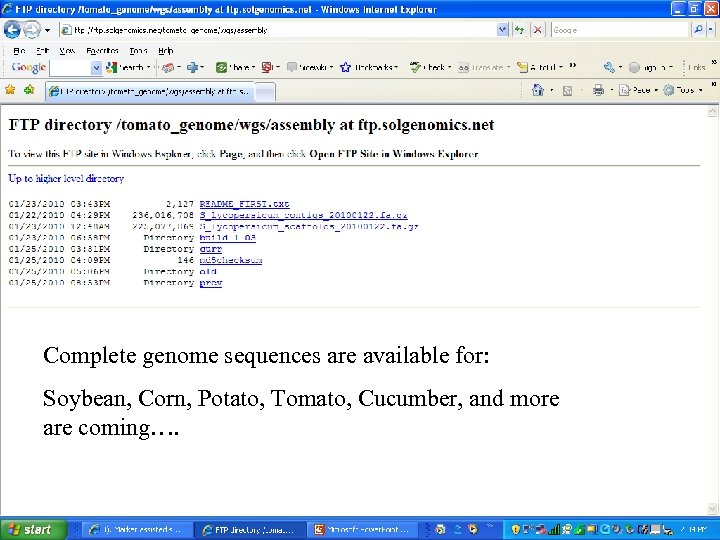
Complete genome sequences are available for: Soybean, Corn, Potato, Tomato, Cucumber, and more are coming….
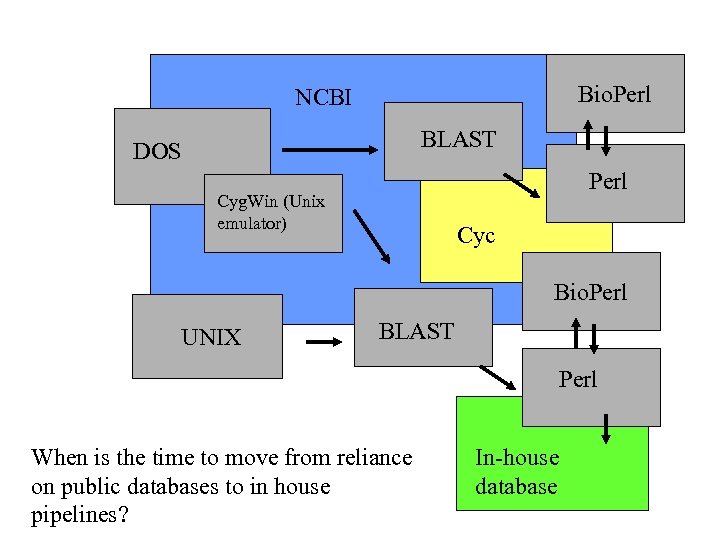
Bio. Perl NCBI BLAST DOS Perl Cyg. Win (Unix emulator) Cyc Bio. Perl UNIX BLAST Perl When is the time to move from reliance on public databases to in house pipelines? In-house database
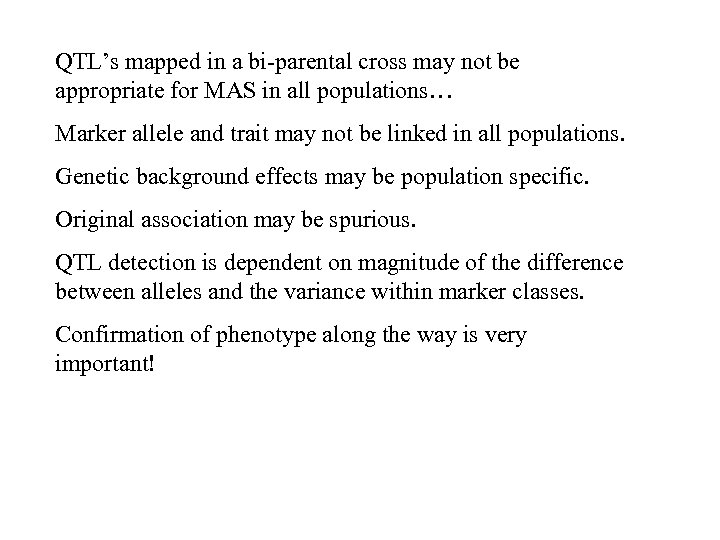
QTL’s mapped in a bi-parental cross may not be appropriate for MAS in all populations… Marker allele and trait may not be linked in all populations. Genetic background effects may be population specific. Original association may be spurious. QTL detection is dependent on magnitude of the difference between alleles and the variance within marker classes. Confirmation of phenotype along the way is very important!

Take home messages: Marker resources exist forward and background selection in elite x elite crosses in tomato. Marker resources are currently not sufficient for QTL discovery in bi-parental or AM populations; they will soon be. The best time to use genetic markers : early generation selection Restructuring of breeding program to integrate markers may include: 1) Increasing genotypic replication (population size) at the expense of replication (consider augmented designs). 2) Collecting objective data.

References: Kaepler, 1997. TAG 95: 618 -621. Frisch, et al. , 1999. Crop Science 39: 1295 -1301. Knapp and Bridges, 1990. Genetics 126: 769 -777. Yu et al. , 2006. Nature Genetics 38: 203 -308. Van Deynze et al. , 2007. BMC Genomics 8: 465 www. biomedcentral. com/content/pdf/1471 -2164 -8 -465. pdf
75361a5d9867b9bba7ac8b46256053fb.ppt