516e5207e9867167111ef7a7c43a0a7c.ppt
- Количество слайдов: 20
 Short comparion GASP ‘ 99 - EGASP ‘ 05 Martin Reese (mreese@omicia. com Omicia Inc. 5980 Horton Street Emeryville, CA 94602 Reese, E-GASP 2005
Short comparion GASP ‘ 99 - EGASP ‘ 05 Martin Reese (mreese@omicia. com Omicia Inc. 5980 Horton Street Emeryville, CA 94602 Reese, E-GASP 2005
 The challenge of annotating a complete eukaryotic genome: A case study in Drosophila melanogaster Martin G. Reese (mgreese@lbl. gov) Nomi L. Harris (nlharris@lbl. gov) George Hartzell (hartzell@cs. berkeley. edu) Suzanna E. Lewis (suzi@fruitfly. berkeley. edu) Later added: Josep April Drosophila Genome Center Department of Molecular and Cell Biology 539 Life Sciences Addition University of California, Berkeley Reese, E-GASP 2005
The challenge of annotating a complete eukaryotic genome: A case study in Drosophila melanogaster Martin G. Reese (mgreese@lbl. gov) Nomi L. Harris (nlharris@lbl. gov) George Hartzell (hartzell@cs. berkeley. edu) Suzanna E. Lewis (suzi@fruitfly. berkeley. edu) Later added: Josep April Drosophila Genome Center Department of Molecular and Cell Biology 539 Life Sciences Addition University of California, Berkeley Reese, E-GASP 2005
 The genome annotation experiment “GASP” 1999 Annotation of 2. 9 Mb of Drosophila melanogaster genomic DNA 44 separate regions Open to everybody, announced on several mailing lists Participants can use any analysis methods they like (gene finding programs, homology searches, by-eye assessment, combination methods, etc. ) and should disclose their methods. “CASP” like 12 participating groups EGASP at least 20 groups 3 Reese, E-GASP 2005
The genome annotation experiment “GASP” 1999 Annotation of 2. 9 Mb of Drosophila melanogaster genomic DNA 44 separate regions Open to everybody, announced on several mailing lists Participants can use any analysis methods they like (gene finding programs, homology searches, by-eye assessment, combination methods, etc. ) and should disclose their methods. “CASP” like 12 participating groups EGASP at least 20 groups 3 Reese, E-GASP 2005
 URL: http: //www-hgc. lbl. gov/homes/reese/genome-annotation 4 Reese, E-GASP 2005
URL: http: //www-hgc. lbl. gov/homes/reese/genome-annotation 4 Reese, E-GASP 2005
 Goals of the experiment Compare and contrast various genome annotation methods Objective assessment of the state of the art in gene finding and functional site prediction Identify outstanding problems in computational methods for the annotation process 5 Reese, E-GASP 2005
Goals of the experiment Compare and contrast various genome annotation methods Objective assessment of the state of the art in gene finding and functional site prediction Identify outstanding problems in computational methods for the annotation process 5 Reese, E-GASP 2005
 Adh contig 2. 9 Mb contiguous Drosophila sequence from the Adh region, one of the best studied genomic regions From chromosome 2 L (34 D-36 A) Ashburner et al. , (to appear in Genetics) 222 gene annotations (as of July 22, 1999) ~450 genes 375, 585 bases are coding (12. 95%) ENCODE region 30 Mb We chose the Adh region because it was thought to be typical. A representative test bed to evaluate annotation techniques. 6 Reese, E-GASP 2005
Adh contig 2. 9 Mb contiguous Drosophila sequence from the Adh region, one of the best studied genomic regions From chromosome 2 L (34 D-36 A) Ashburner et al. , (to appear in Genetics) 222 gene annotations (as of July 22, 1999) ~450 genes 375, 585 bases are coding (12. 95%) ENCODE region 30 Mb We chose the Adh region because it was thought to be typical. A representative test bed to evaluate annotation techniques. 6 Reese, E-GASP 2005
 Adh paper (to appear in Genetics) URL: http: //www. fruitfly. org/publications/PDF/ADH. pdf 7 Reese, E-GASP 2005
Adh paper (to appear in Genetics) URL: http: //www. fruitfly. org/publications/PDF/ADH. pdf 7 Reese, E-GASP 2005
 Submissions “MAGPIE” Team: T. Gaasterland et al. Computational Genomics Group, The Sanger Centre: V. Solovyev University of Erlangen: U. Ohler Genome Annotation Group, The Sanger Centre: E. Birney Oakridge Nat. Laboratory “GRAIL”: R. Mural et al. CBS Technical University of Denmark “HMMGene”: A. Krogh Georgia Institute of Technology “Gene. Mark. hmm”: M. Borodovsky IMIM, Spain “Gene. ID”: Roderic Guigó et al. Fred Hutchinson Cancer Center “BLOCKS”: Henikoff & Henikoff GSF, Neuherberg, Germany” M. Scherf Mount Sinai School of Medicine”: Gary Benson UCB/UC Santa Cruz/Neomorphic “Genie”: M. Reese and D. Kulp 8 Reese, E-GASP 2005
Submissions “MAGPIE” Team: T. Gaasterland et al. Computational Genomics Group, The Sanger Centre: V. Solovyev University of Erlangen: U. Ohler Genome Annotation Group, The Sanger Centre: E. Birney Oakridge Nat. Laboratory “GRAIL”: R. Mural et al. CBS Technical University of Denmark “HMMGene”: A. Krogh Georgia Institute of Technology “Gene. Mark. hmm”: M. Borodovsky IMIM, Spain “Gene. ID”: Roderic Guigó et al. Fred Hutchinson Cancer Center “BLOCKS”: Henikoff & Henikoff GSF, Neuherberg, Germany” M. Scherf Mount Sinai School of Medicine”: Gary Benson UCB/UC Santa Cruz/Neomorphic “Genie”: M. Reese and D. Kulp 8 Reese, E-GASP 2005
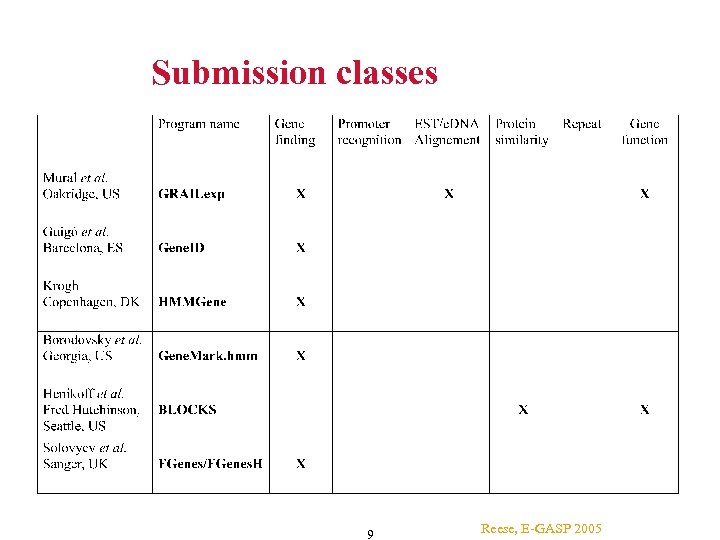 Submission classes 9 Reese, E-GASP 2005
Submission classes 9 Reese, E-GASP 2005
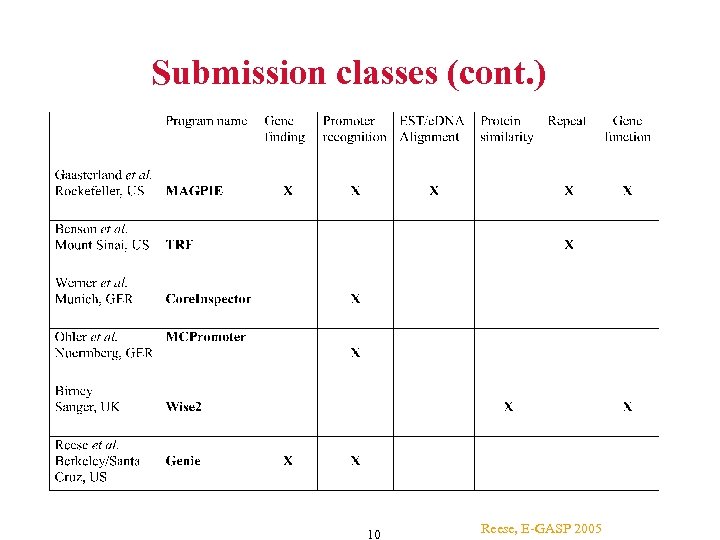 Submission classes (cont. ) 10 Reese, E-GASP 2005
Submission classes (cont. ) 10 Reese, E-GASP 2005
 Measuring success By nucleotide Sensitivity/Specificity (Sn/Sp) By exon Sn/Sp Missed exons (ME), wrong exons (WE) By gene Sn/Sp Missed genes (MG), wrong genes (WG) Average overlap statistics Based on Burset and Guigo (1996), “Evaluation of gene structure prediction programs”. Genomics, 34(3), 353 -367. 11 Reese, E-GASP 2005
Measuring success By nucleotide Sensitivity/Specificity (Sn/Sp) By exon Sn/Sp Missed exons (ME), wrong exons (WE) By gene Sn/Sp Missed genes (MG), wrong genes (WG) Average overlap statistics Based on Burset and Guigo (1996), “Evaluation of gene structure prediction programs”. Genomics, 34(3), 353 -367. 11 Reese, E-GASP 2005
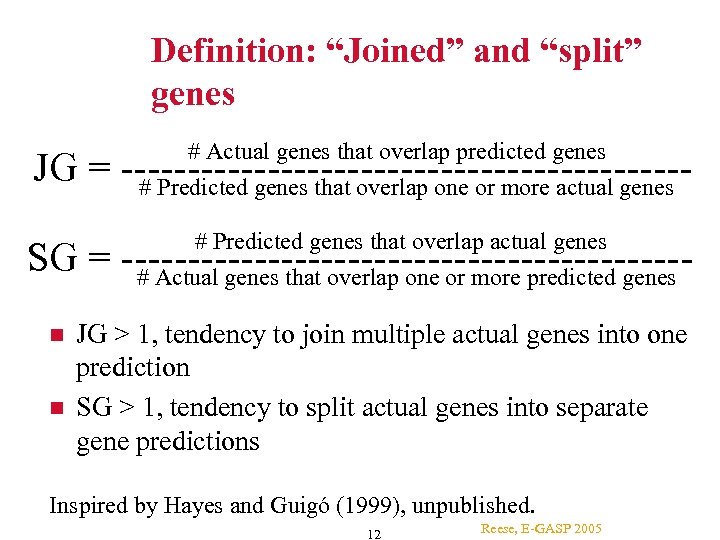 Definition: “Joined” and “split” genes # Actual genes that overlap predicted genes # Predicted genes that overlap one or more actual genes JG = ---------------------# Predicted genes that overlap actual genes # Actual genes that overlap one or more predicted genes SG = --------------------- JG > 1, tendency to join multiple actual genes into one prediction SG > 1, tendency to split actual genes into separate gene predictions Inspired by Hayes and Guigó (1999), unpublished. 12 Reese, E-GASP 2005
Definition: “Joined” and “split” genes # Actual genes that overlap predicted genes # Predicted genes that overlap one or more actual genes JG = ---------------------# Predicted genes that overlap actual genes # Actual genes that overlap one or more predicted genes SG = --------------------- JG > 1, tendency to join multiple actual genes into one prediction SG > 1, tendency to split actual genes into separate gene predictions Inspired by Hayes and Guigó (1999), unpublished. 12 Reese, E-GASP 2005
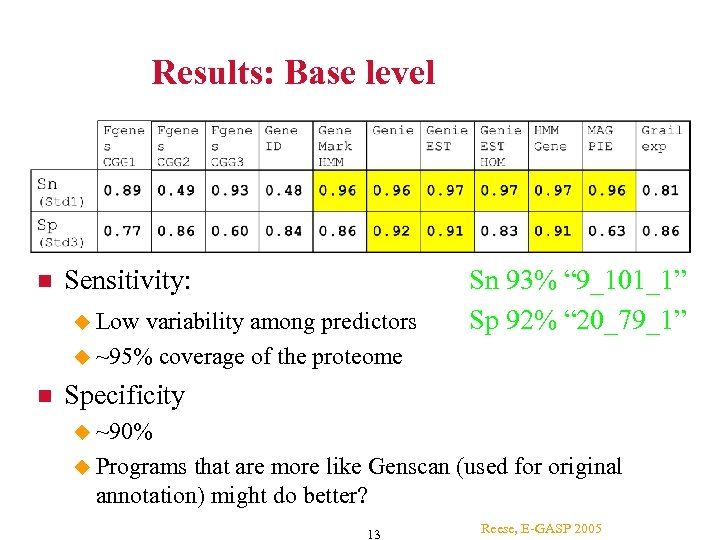 Results: Base level Sensitivity: Low variability among predictors ~95% coverage of the proteome Sn 93% “ 9_101_1” Sp 92% “ 20_79_1” Specificity ~90% Programs that are more like Genscan (used for original annotation) might do better? 13 Reese, E-GASP 2005
Results: Base level Sensitivity: Low variability among predictors ~95% coverage of the proteome Sn 93% “ 9_101_1” Sp 92% “ 20_79_1” Specificity ~90% Programs that are more like Genscan (used for original annotation) might do better? 13 Reese, E-GASP 2005
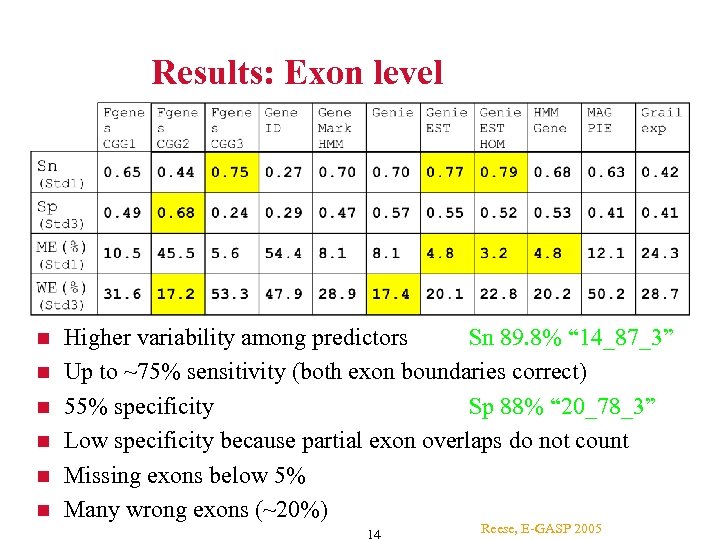 Results: Exon level Higher variability among predictors Sn 89. 8% “ 14_87_3” Up to ~75% sensitivity (both exon boundaries correct) 55% specificity Sp 88% “ 20_78_3” Low specificity because partial exon overlaps do not count Missing exons below 5% Many wrong exons (~20%) 14 Reese, E-GASP 2005
Results: Exon level Higher variability among predictors Sn 89. 8% “ 14_87_3” Up to ~75% sensitivity (both exon boundaries correct) 55% specificity Sp 88% “ 20_78_3” Low specificity because partial exon overlaps do not count Missing exons below 5% Many wrong exons (~20%) 14 Reese, E-GASP 2005
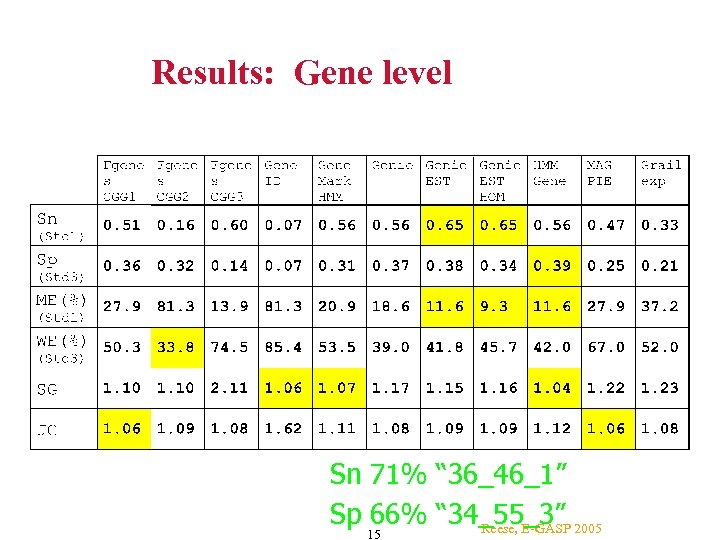 Results: Gene level Sn 71% “ 36_46_1” Sp 66% “ 34_55_3” 2005 Reese, E-GASP 15
Results: Gene level Sn 71% “ 36_46_1” Sp 66% “ 34_55_3” 2005 Reese, E-GASP 15
 Results: Gene level 60% of actual genes predicted completely correct Specificity only 30 -40% 5 -10% missed genes (comparable to Sanger Center) 40% wrong genes, a lot of short genes overpredicted (possibly not annotated in Standard 3) Splitting genes is a bigger problem than joining genes Sn 71% “ 36_46_1” Sp 66% “ 34_55_3” 16 Reese, E-GASP 2005
Results: Gene level 60% of actual genes predicted completely correct Specificity only 30 -40% 5 -10% missed genes (comparable to Sanger Center) 40% wrong genes, a lot of short genes overpredicted (possibly not annotated in Standard 3) Splitting genes is a bigger problem than joining genes Sn 71% “ 36_46_1” Sp 66% “ 34_55_3” 16 Reese, E-GASP 2005
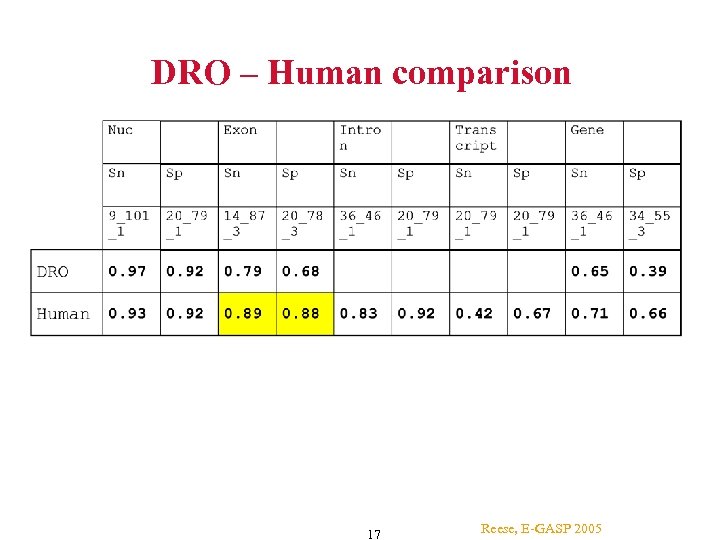 DRO – Human comparison 17 Reese, E-GASP 2005
DRO – Human comparison 17 Reese, E-GASP 2005
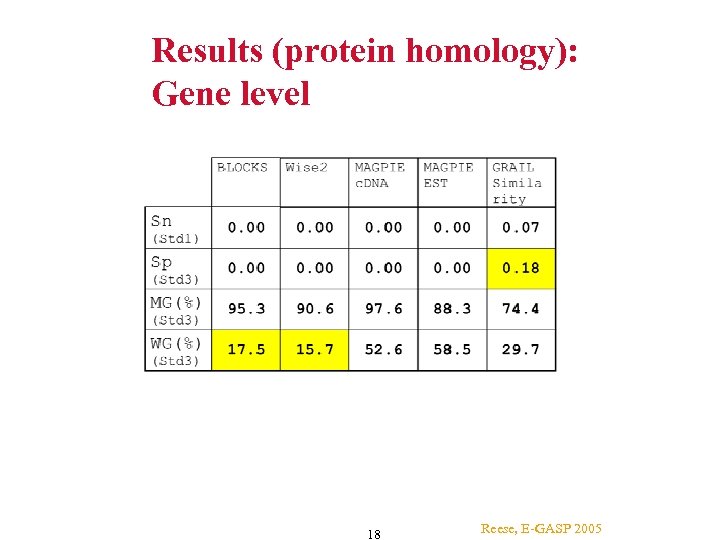 Results (protein homology): Gene level 18 Reese, E-GASP 2005
Results (protein homology): Gene level 18 Reese, E-GASP 2005
 Discussion Good predictive improvements “expression” improves predictions “gene finding” became “automatic annotation” tools Gene sensitivity/specificity at roughly 70% is excellent No correct answer/real golden standard (like CASP) Superb community 19 Reese, E-GASP 2005
Discussion Good predictive improvements “expression” improves predictions “gene finding” became “automatic annotation” tools Gene sensitivity/specificity at roughly 70% is excellent No correct answer/real golden standard (like CASP) Superb community 19 Reese, E-GASP 2005
 Open questions How many protein coding genes/loci missed? How many total human protein coding loci are there? (Dro <14, 500) How much and what is the function of array detected transcripts (coding non-coding? ) Can we get an exhaustive alternative splicing “golden standard”? 20 Reese, E-GASP 2005
Open questions How many protein coding genes/loci missed? How many total human protein coding loci are there? (Dro <14, 500) How much and what is the function of array detected transcripts (coding non-coding? ) Can we get an exhaustive alternative splicing “golden standard”? 20 Reese, E-GASP 2005


