cb2618c56ceece38ab6f0fda00925cbf.ppt
- Количество слайдов: 49
Sequencing the Maize (B 73) Genome Maize Genome Sequencing Consortium Genome Sequencing Center

The Team • WU Genome Sequencing Center (R. Wilson, PI) - Bob Fulton, Pat Minx, Sandy Clifton • Arizona Genome Institute (R. Wing) • Cold Spring Harbor Laboratory - D. Ware, L. Stein - R. Mc. Combie, R. Martienssen • Iowa State University (P. Schnable & S. Aluru) • The Maize research community
The Plan

Progress as of 9/30/06


Agenda 9: 00 – 9: 15 Introductions and Project Overview (Rick Wilson) 9: 15 – 10: 15 Plans and Progress – WU/AGI/CSHL/ISU Project Map and Tile Path Selection (Rod Wing) Library Construction and Production (Lucinda Fulton) Sequence Improvement (Bob Fulton, Dick Mc. Combie, Rod Wing) Data Submission (Joanne Nelson) Annotation and Data Display (Doreen Ware) Outreach (Rick Wilson) 10: 15 - 10: 30 Break 10: 30 – 11: 00 Plans and Progress – DOE Project (Dan Rohksar) 11: 00 – 11: 30 Future Plans and Collaborations Pat Schnable (by phone) - retrotransposons 11: 30 – Noon Executive Session Noon – 1: 00 Working Lunch and Discussion 1: 00 Depart for Airport
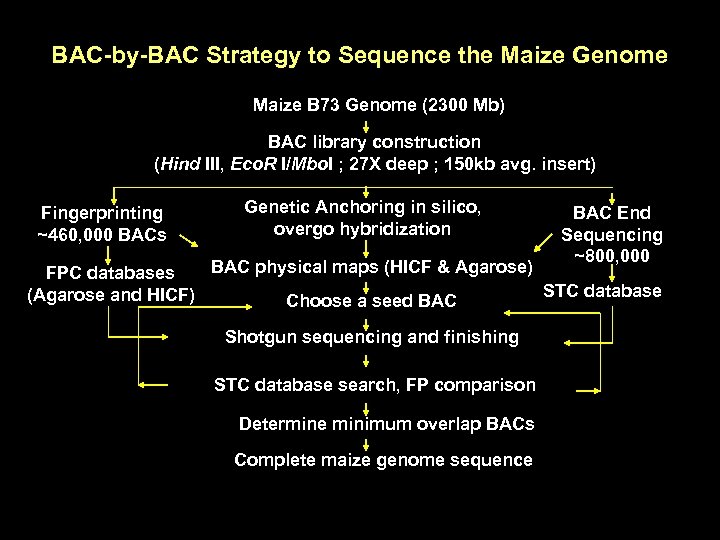
BAC-by-BAC Strategy to Sequence the Maize Genome Maize B 73 Genome (2300 Mb) BAC library construction (Hind III, Eco. R I/Mbo. I ; 27 X deep ; 150 kb avg. insert) Fingerprinting ~460, 000 BACs Genetic Anchoring in silico, overgo hybridization BAC End Sequencing ~800, 000 BAC physical maps (HICF & Agarose) FPC databases STC database (Agarose and HICF) Choose a seed BAC Shotgun sequencing and finishing STC database search, FP comparison Determine minimum overlap BACs Complete maize genome sequence

Map Summary 1. Total Assembled Contigs: 721 – Equal to 2, 150 Mb, 93. 5% coverage of 2300 Mb genome – Anchored: 421 ctgs, 86. 1% the genome – average anchored contig size: 4. 7 Mb – Unanchored: 300 ctgs, 7. 4% coverage average unanchored contig size: 0. 56 Mb – 189 of the 300 unanchored contigs are less than 10 clones – Largest anchored contig 22. 9 Mb in Chr 9 – Largest unanchored contig 6. 7 Mb 2. Total FPC Markers: – STS markers: – Overgo Markers: – Anchored markers: 25, 924 9, 129 14, 877 1918

MTP Selection • Seed BACs: 4000, done • Mega Contig: 197, done • Clone Walking from Seed BACs: 2, 800 done; in progress • Total clones picked = 6, 997 • On track to deliver 1000 clones/month until maze MTP is complete
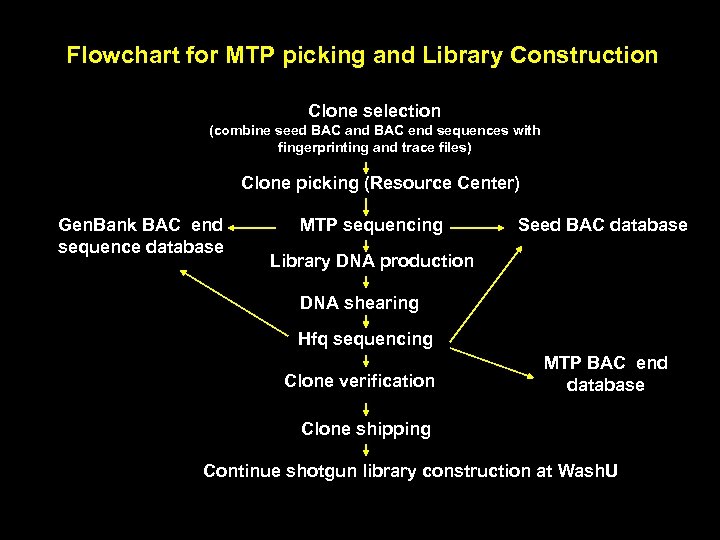
Flowchart for MTP picking and Library Construction Clone selection (combine seed BAC and BAC end sequences with fingerprinting and trace files) Clone picking (Resource Center) Gen. Bank BAC end sequence database MTP sequencing Seed BAC database Library DNA production DNA shearing Hfq sequencing Clone verification MTP BAC end database Clone shipping Continue shotgun library construction at Wash. U
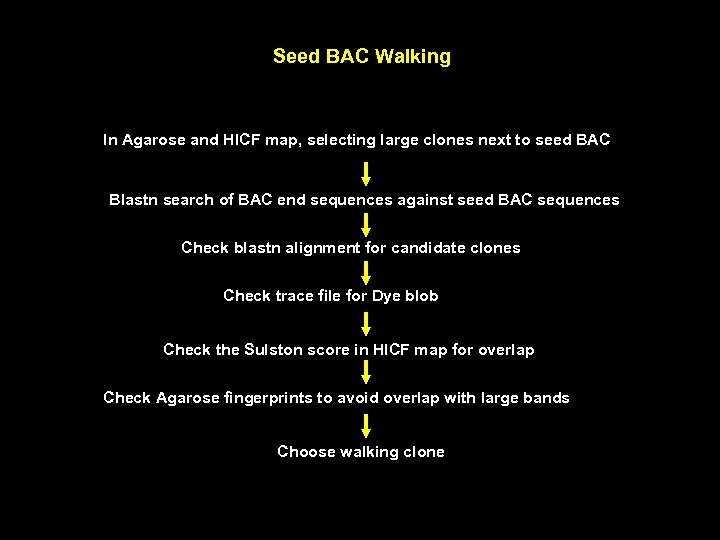
Seed BAC Walking In Agarose and HICF map, selecting large clones next to seed BAC Blastn search of BAC end sequences against seed BAC sequences Check blastn alignment for candidate clones Check trace file for Dye blob Check the Sulston score in HICF map for overlap Check Agarose fingerprints to avoid overlap with large bands Choose walking clone
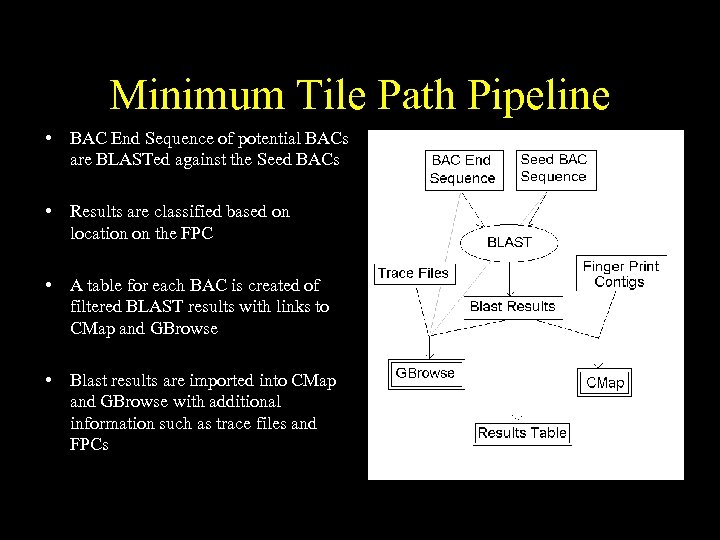
Minimum Tile Path Pipeline • BAC End Sequence of potential BACs are BLASTed against the Seed BACs • Results are classified based on location on the FPC • A table for each BAC is created of filtered BLAST results with links to CMap and GBrowse • Blast results are imported into CMap and GBrowse with additional information such as trace files and FPCs
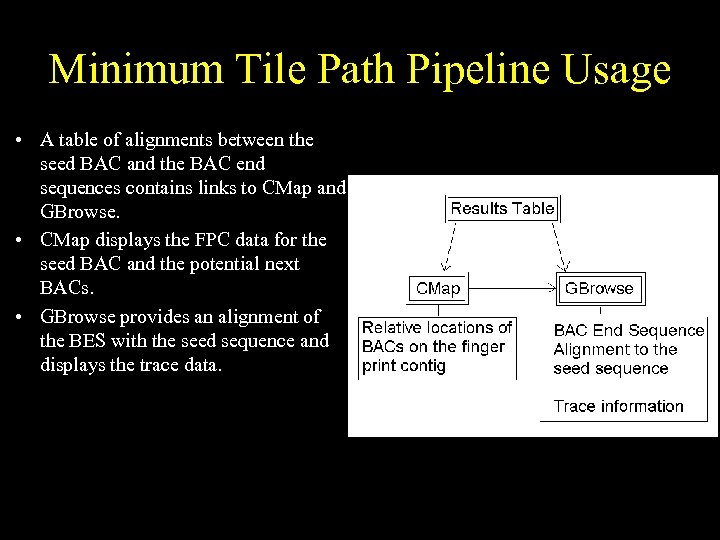
Minimum Tile Path Pipeline Usage • A table of alignments between the seed BAC and the BAC end sequences contains links to CMap and GBrowse. • CMap displays the FPC data for the seed BAC and the potential next BACs. • GBrowse provides an alignment of the BES with the seed sequence and displays the trace data.
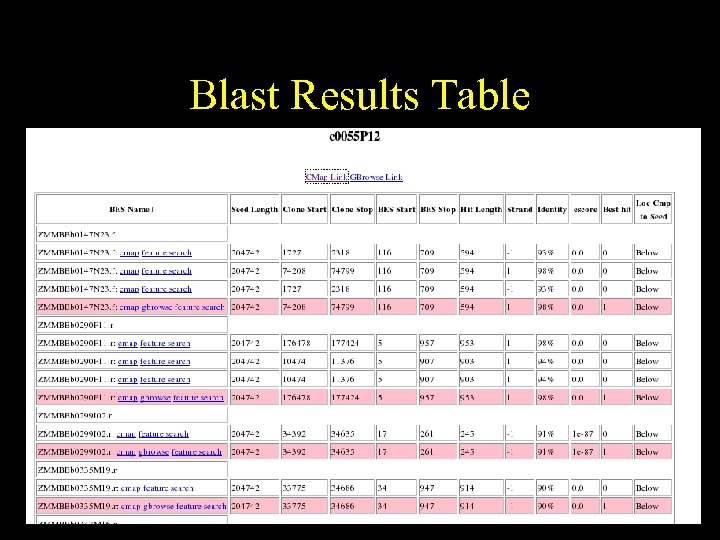
Blast Results Table

Maize Production Sequencing v. Shotgun of 19, 000 BACs v. Fosmid End Sequencing of 1 Million Reads v. BAC End Sequencing of 220, 000 clones

Maize BAC shotgun BAC DNA received from AGI or prepared at the GSC Small Scale Library Construction Production Sequencing - 1, 536 reads/project Automated Shotgun_done
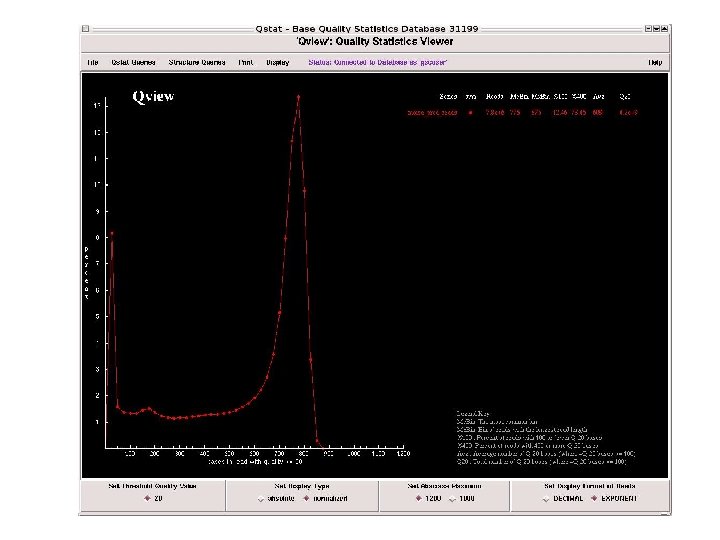

To date 3, 106 BAC clones are shotgun_done
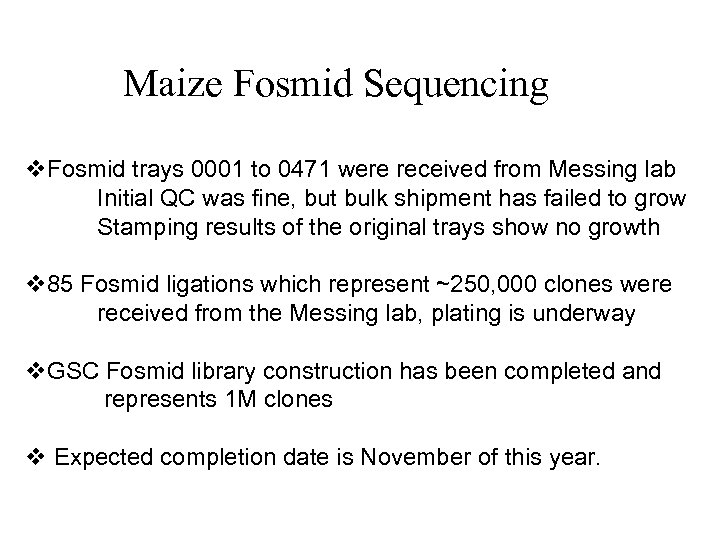
Maize Fosmid Sequencing v. Fosmid trays 0001 to 0471 were received from Messing lab Initial QC was fine, but bulk shipment has failed to grow Stamping results of the original trays show no growth v 85 Fosmid ligations which represent ~250, 000 clones were received from the Messing lab, plating is underway v. GSC Fosmid library construction has been completed and represents 1 M clones v Expected completion date is November of this year.
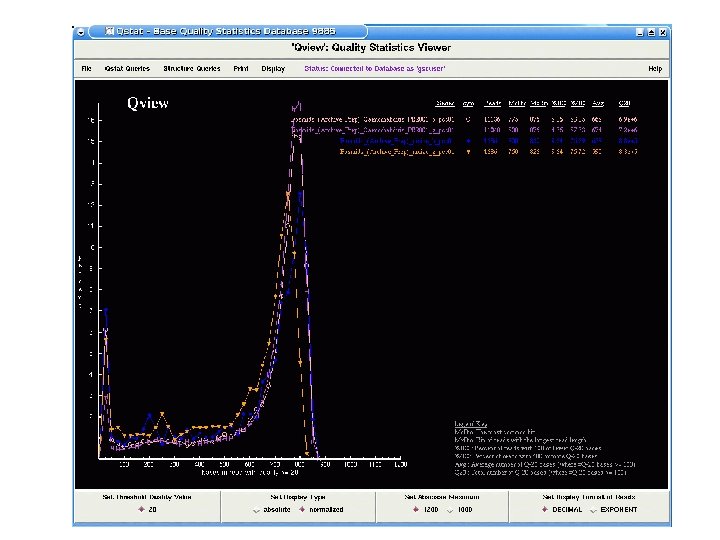
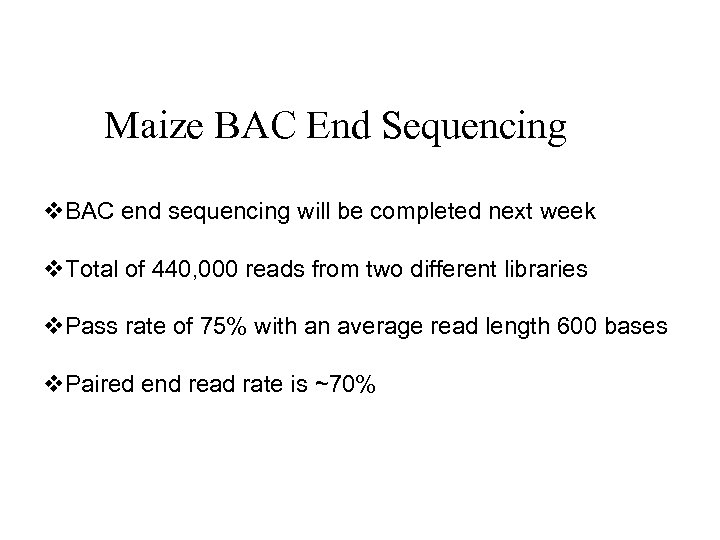
Maize BAC End Sequencing v. BAC end sequencing will be completed next week v. Total of 440, 000 reads from two different libraries v. Pass rate of 75% with an average read length 600 bases v. Paired end read rate is ~70%
Sequence Improvement Pipeline • Shotgun_done triggers the prefinishing pipeline • Initial identification of “do finish” regions • Manual sorting and use of autoedit(Gordon) to break apart misassembly. • Autofinish(Gordon) used to choose directed reactions for all gaps and regions of low quality in “do finish” regions • Reassembly and 2 nd iteration of prefinishing pipeline • Final identification of “do finish” regions and handoff to finishing pipeline

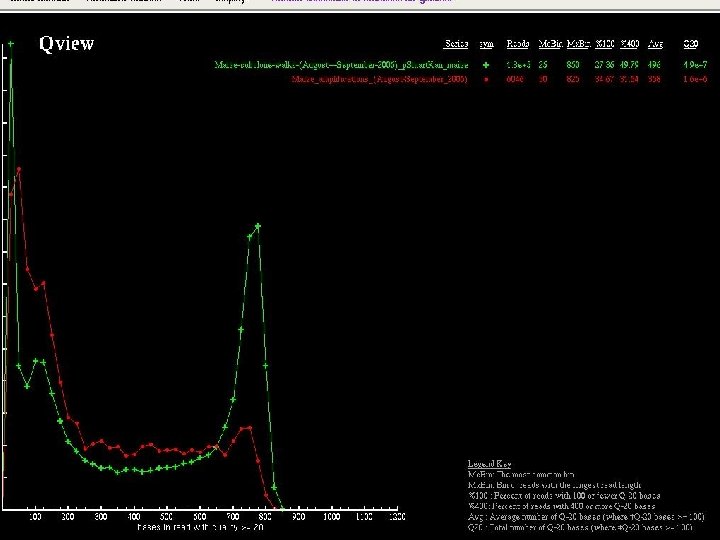
Clone Improvement through the Prefinishing Pipeline
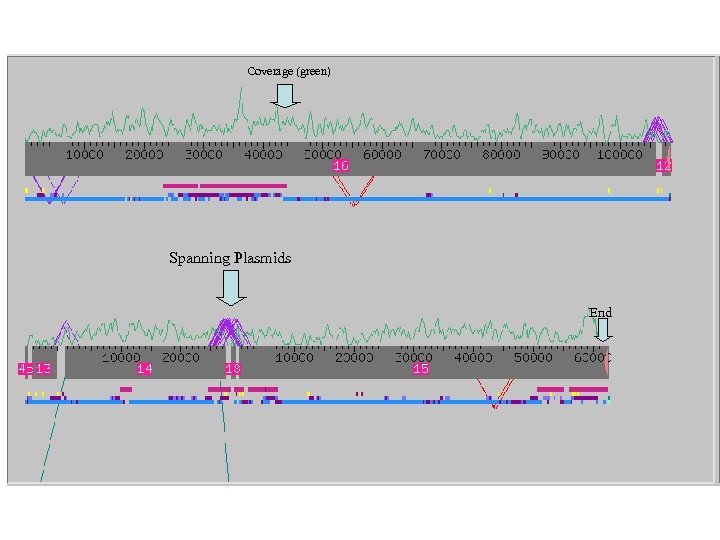
Coverage (green) Spanning Plasmids End
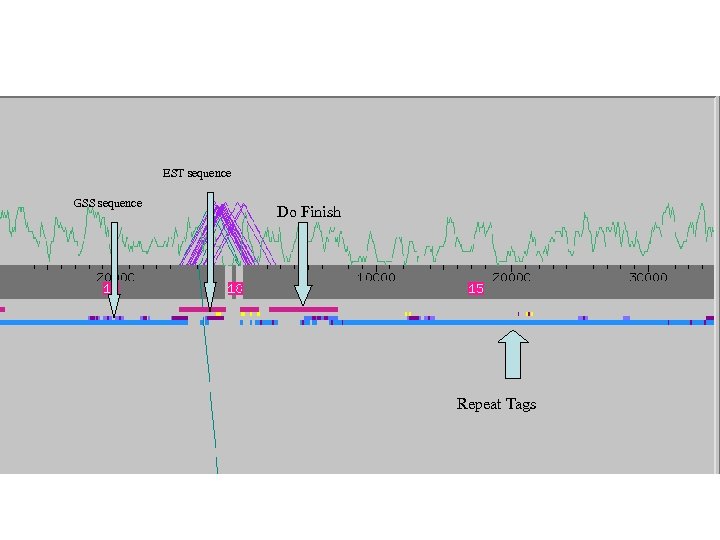
EST sequence GSS sequence Do Finish Repeat Tags
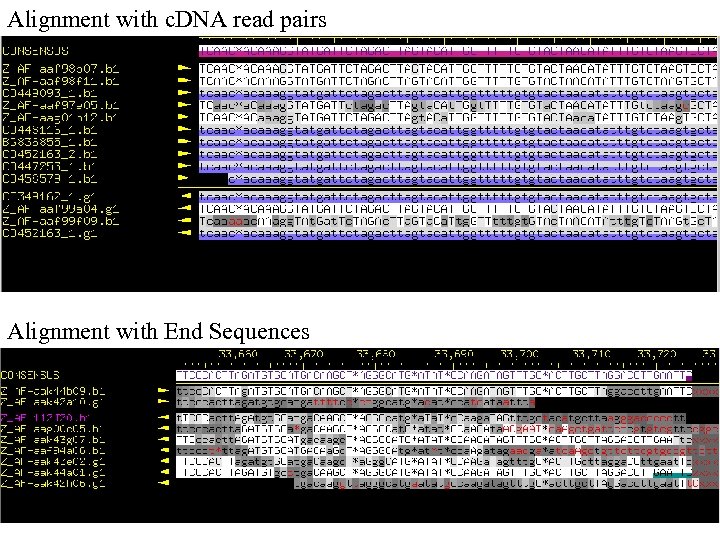
Alignment with c. DNA read pairs Alignment with End Sequences

Future Plans for Improved Throughput • Automated Shotgun-done status assigning • Overlap Evaluation at Prefinishing • Addition of Fosmid End Pairs at Prefinishing • Direct Sequencing for Unspanned Gaps • Additional Finishing Staff Hired at all 3 Centers

Maize clone submissions clone status shotgun complete 2 rounds of prefinish in finishing finished submission keywords HTGS_PHASE 1; HTGS_FULLTOP HTGS_PHASE 1; HTGS_PREFIN HTGS_PHASE 1; HTGS_ACTIVEFIN HTGS_PHASE 1; HTGS_IMPROVED Query Gen. Bank by keywords zea mays[ORGN] AND HTGS_PREFIN[KYWD] AND WUGSC[CNTR] zea mays[ORGN] AND HTGS_IMPROVED[KYWD] AND WUGSC[CNTR] Restrict by date range: zea mays[ORGN] AND WUGSC[CNTR] AND HTGS_FULLTOP[KYWD] AND 2006/09[PDAT] zea mays[ORGN] AND WUGSC[CNTR] AND HTGS_FULLTOP[KYWD] AND 2006/09/26: 2006/10/03[PDAT]

HTGS_IMPROVED submissions Pick a clonename, any clonename DEFINITION Zea mays chromosome 4 clone CH 201 -11 H 16; ZMMBBc 0011 H 16 Center project name: Z_AF-11 H 16 Improved sequence is annotated on submission record Where possible, contigs have been ordered and oriented based on read pairing. and these regions are designated as scaffolds. Small contigs (<2 kb) that don’t represent a clone end, don’t contain improved sequence, or are not part of a scaffold are removed from the final submission. Contigs are screened for bacterial contamination

FEATURES source Location/Qualifiers 1. . 173904 /organism="Zea mays" /mol_type="genomic DNA" /db_xref="taxon: 4577" /chromosome="unknown" /clone="CH 201 -112 C 8; ZMMBBc 0112 C 08" misc_feature 1. . 51940 /note="scaffold_name: Scaffold 1" misc_feature 1. . 36440 /note="assembly_name: Contig 245 clone_end: left vector_side: T 7" gap 36441. . 36540 /estimated_length=unknown misc_feature 36541. . 51940 /note="assembly_name: Contig 240" misc_feature 51941. . 129231 /note="scaffold_name: Scaffold 2" gap 51941. . 52040 /estimated_length=unknown misc_feature 52041. . 59371 /note="assembly_name: Contig 250”. . . misc_feature 120342. . 122491 /note="Improved sequence. " misc_feature 128142. . 129231 /note="Improved sequence. " misc_feature 129232. . 139656 /note="scaffold_name: Scaffold 3". . .

Gen. Bank 1005 254 1532 357 HTGS_FULLTOP PREFIN_DONE ACTIVE_FIN HTGS_IMPROVED

Ongoing work at CSHL • • • BAC Annotations Levels Data Analysis Display Project Management Collaborations
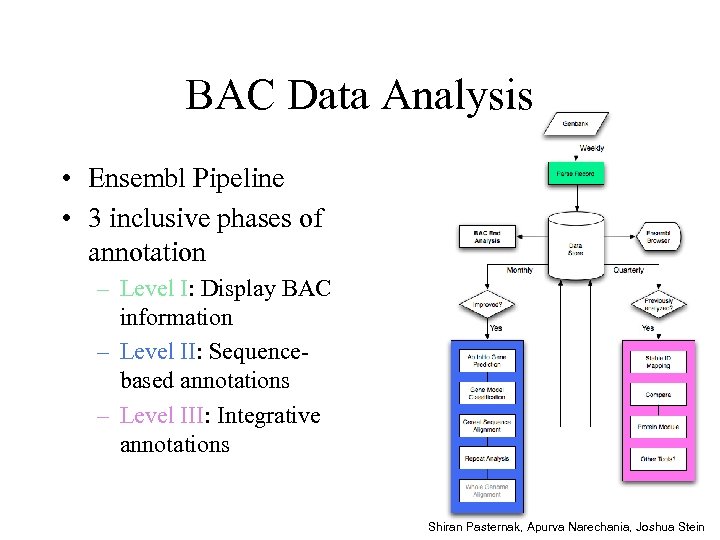
BAC Data Analysis • Ensembl Pipeline • 3 inclusive phases of annotation – Level I: Display BAC information – Level II: Sequencebased annotations – Level III: Integrative annotations Shiran Pasternak, Apurva Narechania, Joshua Stein
Application of Mathematical Repeat Analysis • Identifies novel repeats w/o dependence on curation. – Based on frequency of 20 -mers in JGI WGS sequence • • Correlates with presence of retroelements. Can modulate threshold to optimize application. Apurva Narechania, Joshua Stein
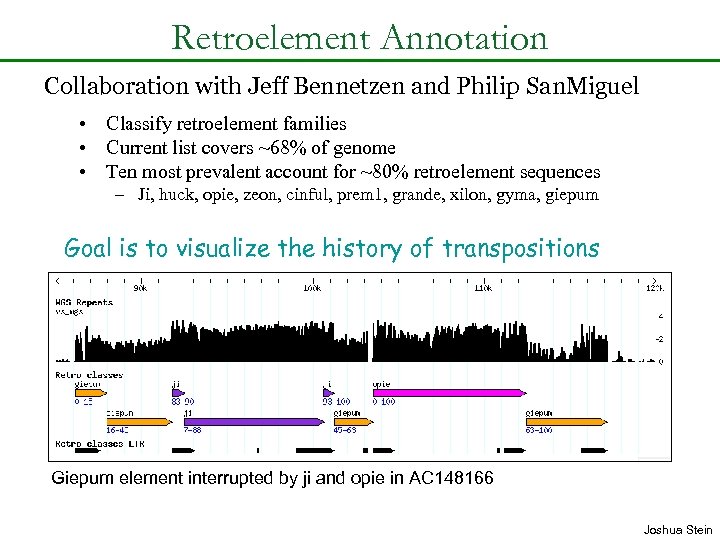
Retroelement Annotation Collaboration with Jeff Bennetzen and Philip San. Miguel • Classify retroelement families • Current list covers ~68% of genome • Ten most prevalent account for ~80% retroelement sequences – Ji, huck, opie, zeon, cinful, prem 1, grande, xilon, gyma, giepum Goal is to visualize the history of transpositions Giepum element interrupted by ji and opie in AC 148166 Joshua Stein
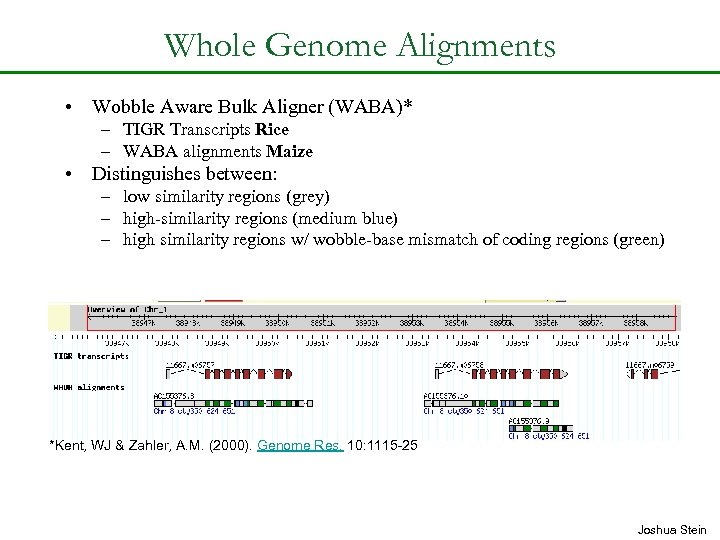
Whole Genome Alignments • Wobble Aware Bulk Aligner (WABA)* – TIGR Transcripts Rice – WABA alignments Maize • Distinguishes between: – low similarity regions (grey) – high-similarity regions (medium blue) – high similarity regions w/ wobble-base mismatch of coding regions (green) *Kent, WJ & Zahler, A. M. (2000). Genome Res. 10: 1115 -25 Joshua Stein
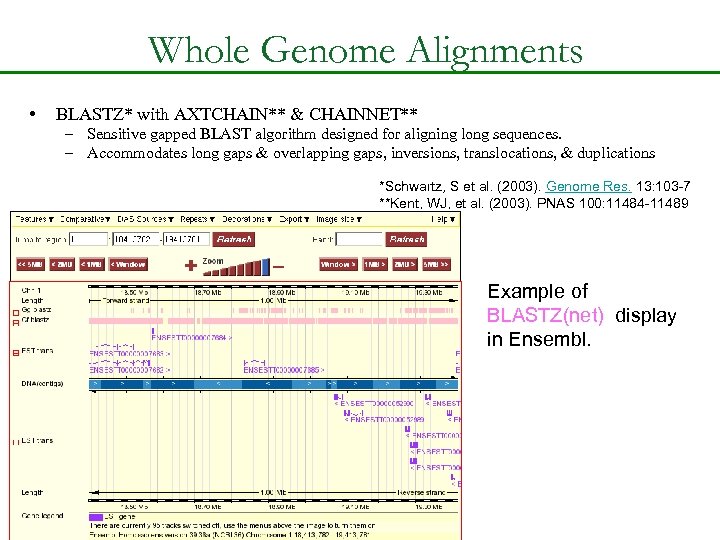
Whole Genome Alignments • BLASTZ* with AXTCHAIN** & CHAINNET** – Sensitive gapped BLAST algorithm designed for aligning long sequences. – Accommodates long gaps & overlapping gaps, inversions, translocations, & duplications *Schwartz, S et al. (2003). Genome Res. 13: 103 -7 **Kent, WJ, et al. (2003). PNAS 100: 11484 -11489 Example of BLASTZ(net) display in Ensembl.

www. maizesequence. org Sequenced BAC FPC Contig Virtual Bin Core Bin Marker Chromosome Synteny Views Main Navigation bar is accessible from every page Contains multiple entry points to the genome
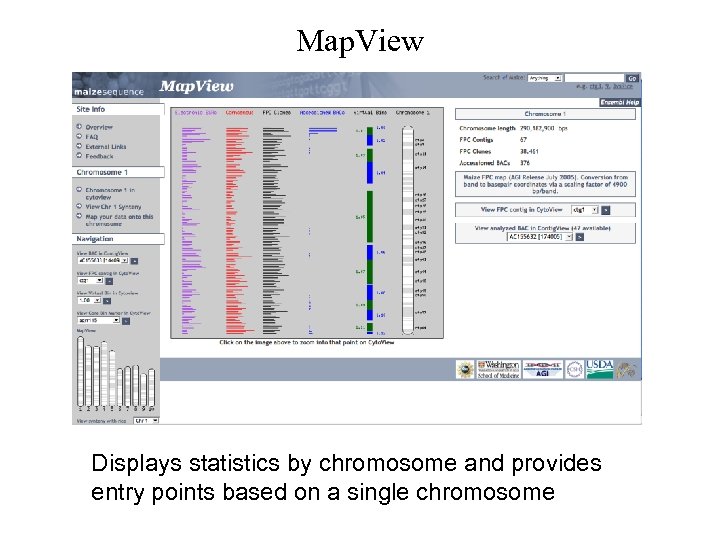
Map. View Displays statistics by chromosome and provides entry points based on a single chromosome
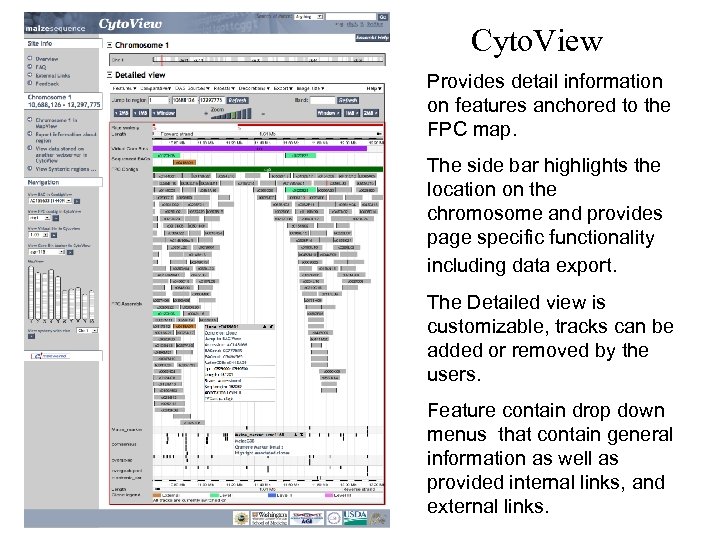
Cyto. View Provides detail information on features anchored to the FPC map. The side bar highlights the location on the chromosome and provides page specific functionality including data export. The Detailed view is customizable, tracks can be added or removed by the users. Feature contain drop down menus that contain general information as well as provided internal links, and external links.
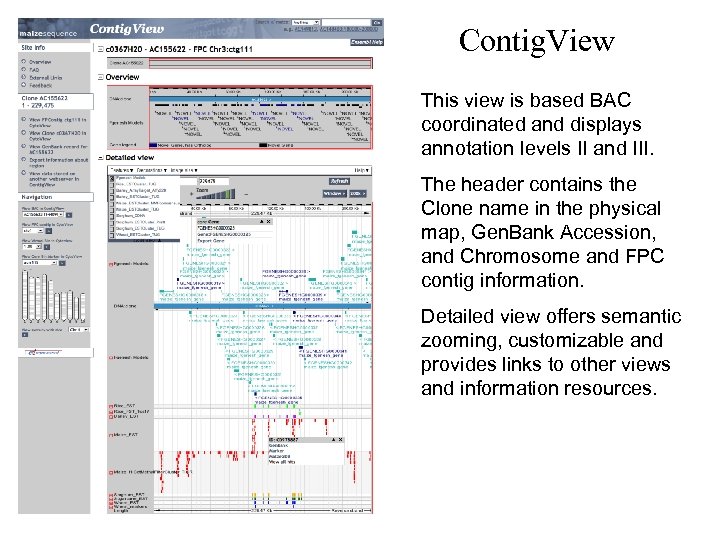
Contig. View This view is based BAC coordinated and displays annotation levels II and III. The header contains the Clone name in the physical map, Gen. Bank Accession, and Chromosome and FPC contig information. Detailed view offers semantic zooming, customizable and provides links to other views and information resources.
Synteny. View

Upcoming Features • Release – October 2006 • Blast. View – December 2006 • BAC Annotation – Level II January, 2007 – Level III annotation April, 2007 – WG alignments June, 2007 • Bio. Mart – January, 2007 • NSF collaborations – Twin. Scan annotations: March, 2007 – Maize Optical Map: July, 2007 – Full-length c. DNAs: December, 2007 • Notification System – Users are notified • When a region of interest is updated • When markers are aligned to a specific sequence – January, 2007
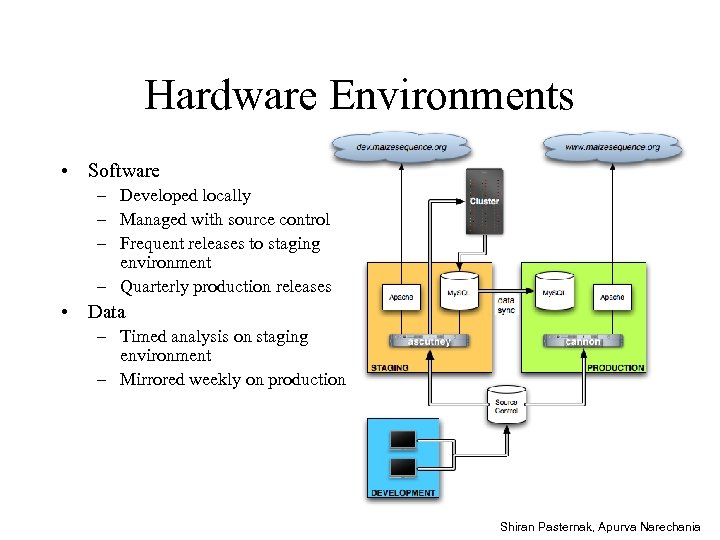
Hardware Environments • Software – Developed locally – Managed with source control – Frequent releases to staging environment – Quarterly production releases • Data – Timed analysis on staging environment – Mirrored weekly on production Shiran Pasternak, Apurva Narechania

Quality Assurance • Unit-testing framework – Binary assertions – Failure report and automatic notification • Software Quality Control – e. g. , code retrieves correct data from the database • Data Quality Control – e. g. , clone in Genbank record exists in FPC map Shiran Pasternak
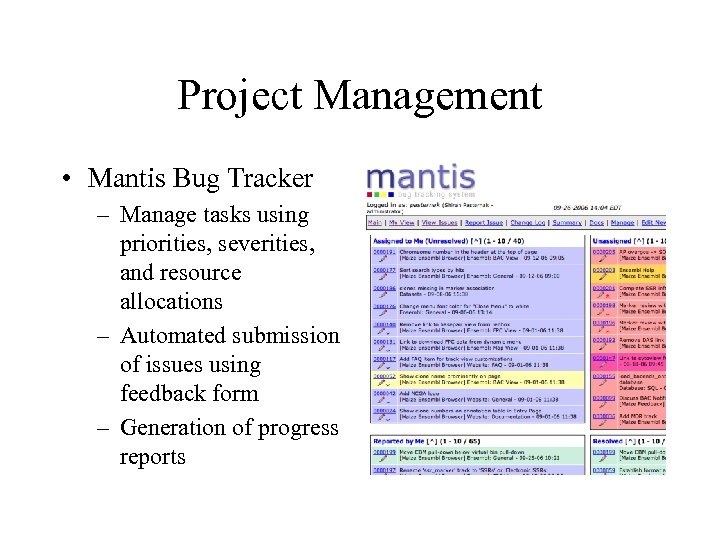
Project Management • Mantis Bug Tracker – Manage tasks using priorities, severities, and resource allocations – Automated submission of issues using feedback form – Generation of progress reports
Project Management • Wiki – Enhances group communication – Meeting notes, flowcharts, specification documents – Maintains history of specifications and design decisions – Seamless editing

Collaborations • • Maize. GDB (Iowa State University, University of Missouri) – C. Lawrence Maize Optical Map (University of Wisconsin) – D. Schwartz Maize Transposon Annotation (University of Georgia, Purdue) – J. Bennetzen, P. San Miguel Ensembl (EBI) – E. Birney Vmatch for Mathematical Repeats (University of Hamburg) – S. Kurtz Maize Full Length c. DNA project (Arizona Genomics Institute) – Y. Yu Twin. Scan (Danforth Plant Science Center) – B. Barbazuk
cb2618c56ceece38ab6f0fda00925cbf.ppt