29abbe9a5219a7b08e8f8b93571dc640.ppt
- Количество слайдов: 66
![Sequence databases and retrieval systems Guy Perrière [ replaced by Manolo Gouy ] Pôle Sequence databases and retrieval systems Guy Perrière [ replaced by Manolo Gouy ] Pôle](https://present5.com/presentation/29abbe9a5219a7b08e8f8b93571dc640/image-1.jpg)
Sequence databases and retrieval systems Guy Perrière [ replaced by Manolo Gouy ] Pôle Bio-Informatique Lyonnais Laboratoire de Biométrie et Biologie Évolutive UMR CNRS n° 5558 Université Claude Bernard – Lyon 1

In the beginning First paper compilation in 1965 (Atlas of Protein Sequences). Development of real databanks at the beginning of the 80’s: Fast access. Make possible analyses that require a lot of data: – Codon usage. – Molecular phylogeny.
General databanks Nucleotide sequences: EMBL/Gen. Bank/DDBJ. Protein sequences: Simple translations of coding regions: – Gen. Pept (from Gen. Bank). – Tr. EMBL (from EMBL). Systems containing additional data: – SWISS-PROT. – PIR.
EMBL Created in 1980 at the European Molecular Biology Laboratory in Heidelberg. Maintained since 1994 at the European Bioinformatics Institute (EBI) near Cambridge. Web server: http: //www. ebi. ac. uk/embl

Gen. Bank Set up in 1979 at the Los Alamos National Laboratory in New Mexico, US. Maintained since 1992 at the National Center for Biotechnology Information (NCBI) in Bethesda. Web server: http: //www. ncbi. nlm. nih. gov/Genbank/index. html

DDBJ Active since 1984 at the National Institute of Genetics (NIG) in Mishima, Japan. Web server: http: //www. ddbj. nig. ac. jp
EMBL / Gen. Bank / DDBJ The International Nucleotide Sequence Database Collaboration : EMBL / Gen. Bank / DDBJ New sequences are exchanged daily between the three centers : --> the three banks have an identical content. Data mainly provided by direct submissions from the authors through Internet: Web forms. Email.
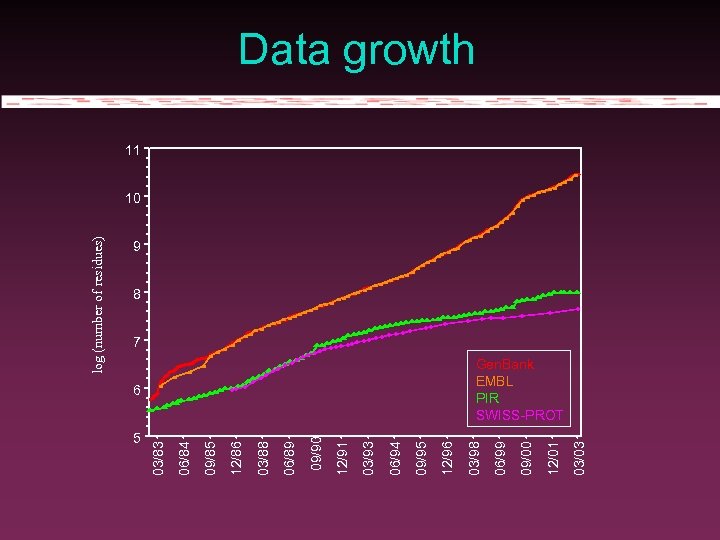
5 03/03 12/01 09/00 06/99 6 03/98 12/96 09/95 06/94 03/93 12/91 09/90 06/89 03/88 12/86 09/85 06/84 03/83 log (number of residues) Data growth 11 10 9 8 7 Gen. Bank EMBL PIR SWISS-PROT
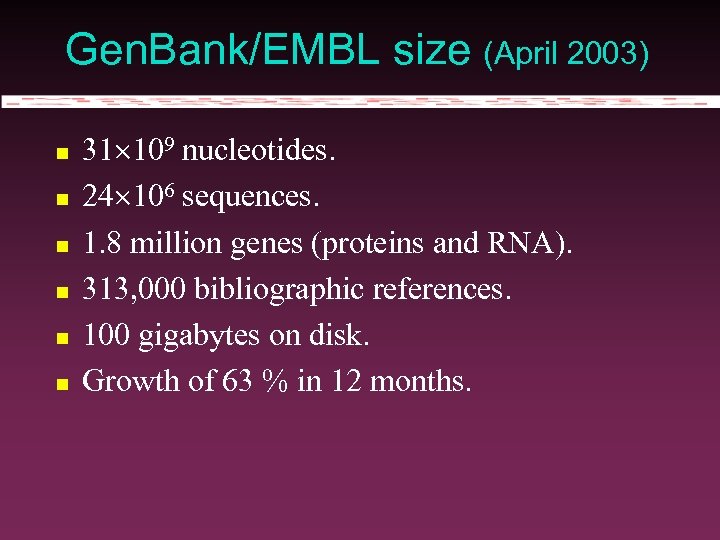
Gen. Bank/EMBL size (April 2003) 31 109 nucleotides. 24 106 sequences. 1. 8 million genes (proteins and RNA). 313, 000 bibliographic references. 100 gigabytes on disk. Growth of 63 % in 12 months.

Taxonomic sampling (April 2003) There are 135, 560 species for which at least one sequence is available. Nine species (0. 007 %) correspond to 62 % of the total. 77, 900 species are represented by only one sequence! Homo sapiens Mus musculus Zea mays Rattus norvegicus Brassica oleracea Arabidopsis thaliana Danio rerio Drosophila melanogaster Oryza sativa 27. 3% 20. 1% 3. 0 % 2. 9 % 2. 3 % 2. 0 % 1. 4 % 0. 9 % The nine most represented species in Gen. Bank/EMBL

Distribution format The banks are distributed as a set of text files called divisions ( 292 for EMBL). A division contains sequences related to: A taxon (e. g. , bacteria, invertebrates, mammals). A class of sequences (EST, HTG, GSS). Within a division, each sequence is called an entry.

Entry structure Information is introduced in structured fields. The format differs in its form between EMBL and Gen. Bank/DDBJ … but not in substance.
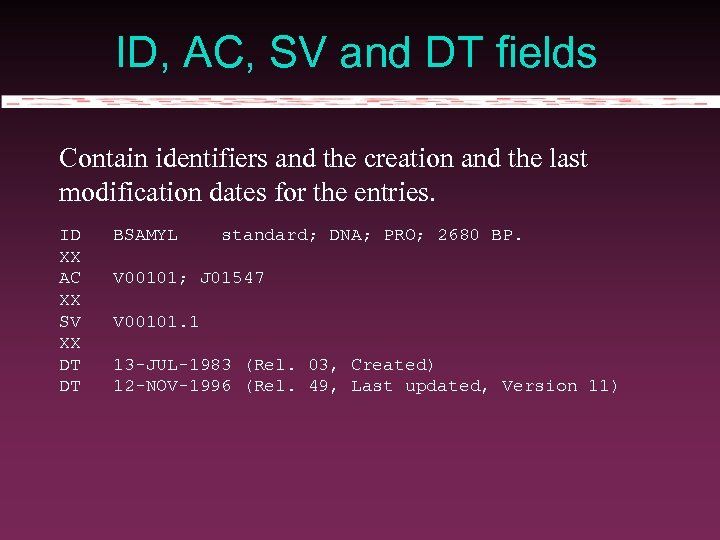
ID, AC, SV and DT fields Contain identifiers and the creation and the last modification dates for the entries. ID XX AC XX SV XX DT DT BSAMYL standard; DNA; PRO; 2680 BP. V 00101; J 01547 V 00101. 1 13 -JUL-1983 (Rel. 03, Created) 12 -NOV-1996 (Rel. 49, Last updated, Version 11)
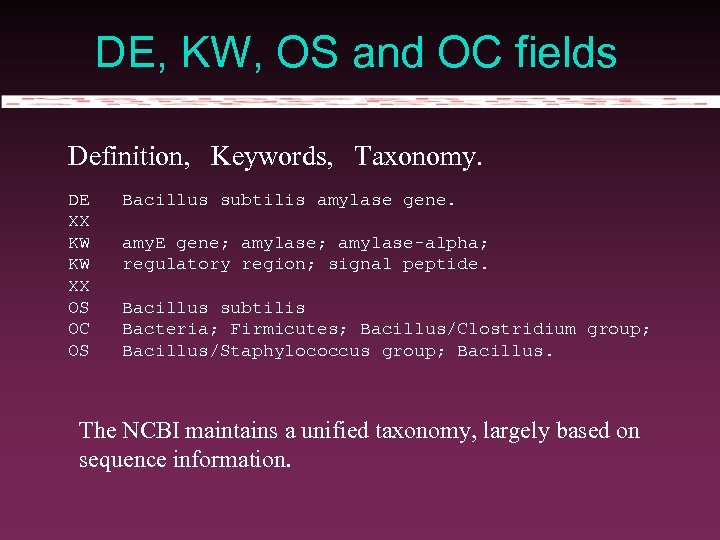
DE, KW, OS and OC fields Definition, Keywords, Taxonomy. DE XX KW KW XX OS OC OS Bacillus subtilis amylase gene. amy. E gene; amylase-alpha; regulatory region; signal peptide. Bacillus subtilis Bacteria; Firmicutes; Bacillus/Clostridium group; Bacillus/Staphylococcus group; Bacillus. The NCBI maintains a unified taxonomy, largely based on sequence information.

RN, RX, RA and RT fields contain bibliographic information. RN RP RX RA RT RT RL … [1] 1 -2680 MEDLINE; 83143299. Yang M. , Galizzi, A. , Henner, D. J. ; "Nucleotide sequence of the amylase gene from Bacillus subtilis"; Nucleic Acids Res. 11: 237 -249(1983).
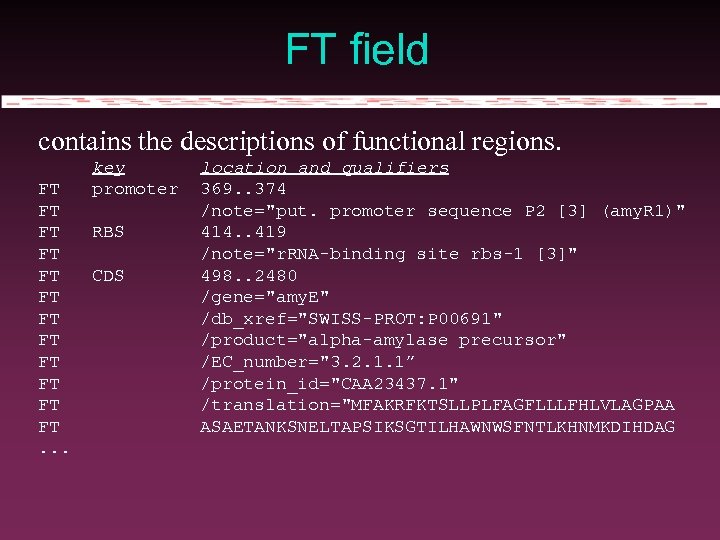
FT field contains the descriptions of functional regions. FT FT FT. . . key promoter RBS CDS location and qualifiers 369. . 374 /note="put. promoter sequence P 2 [3] (amy. R 1)" 414. . 419 /note="r. RNA-binding site rbs-1 [3]" 498. . 2480 /gene="amy. E" /db_xref="SWISS-PROT: P 00691" /product="alpha-amylase precursor" /EC_number="3. 2. 1. 1” /protein_id="CAA 23437. 1" /translation="MFAKRFKTSLLPLFAGFLLLFHLVLAGPAA ASAETANKSNELTAPSIKSGTILHAWNWSFNTLKHNMKDIHDAG
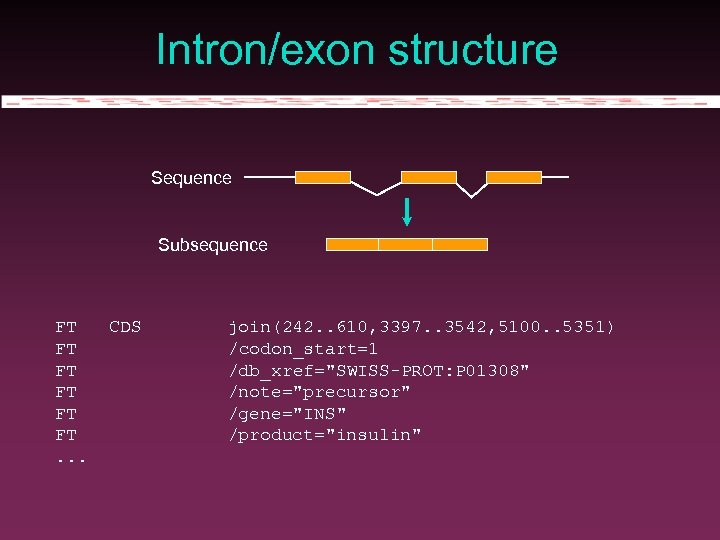
Intron/exon structure Sequence Subsequence FT FT FT. . . CDS join(242. . 610, 3397. . 3542, 5100. . 5351) /codon_start=1 /db_xref="SWISS-PROT: P 01308" /note="precursor" /gene="INS" /product="insulin"

SQ field Contains the sequence iself SQ // Sequence 2680 BP; 825 gctcatgccg agaatagaca agaatcaatt gcttgcgcct ccatacattc ttcgcttggc gtttctgctt cggtatgtga (. . . ) gatggtttct tttttgttca tgttgcacaa tataaatgtg cctgcaagga tgctgatatt A; 520 C; 642 G; 693 T; 0 other; ccaaagaaga actgtaaaaa cgggtgaagc ttgcggtagt ggtgcttacg atgtacgaca tgaaaatgat tcttcttttt atcgtctgcg ttgtgaagct ggcttacaga agagcggtaa agcagcgaat gggggattcc gcggcgttct aagaagaaat 60 120 180 240 taaatcagac aaaacttttc tcttgcaaaa gtttgtgaag aaatacttca caaaaa gacatcaaag agaaacatac gtctgcattt gcgccggagc 2580 2640 2680

Errors in databanks There a lot of errors in the nucleotide sequence databanks: In annotations: – Inaccuracies, omissions, and even mistakes. – Inconsistencies between entries. In the sequences themselves: – Sequencing errors. – Cloning vectors inserted.
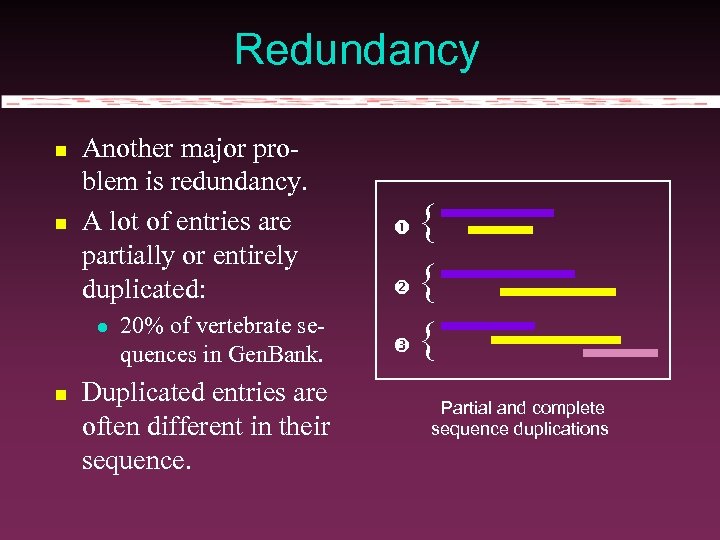
Redundancy Another major problem is redundancy. A lot of entries are partially or entirely duplicated: 20% of vertebrate sequences in Gen. Bank. Duplicated entries are often different in their sequence. { { { Partial and complete sequence duplications
Protein sequence databases Translation of Coding DNA Sequences (CDS) from EMBL/Gen. Bank/DDBJ. Consultation of publications or patents. Very small number of direct protein sequence submission by authors. In Swiss. Prot and PIR: additional annotations.

SWISS-PROT Created by Amos Bairoch in 1986 at the Department of Medical Biochemistry in Geneva. Maintained by the Swiss Institute of Bioinformatics (SIB) and funded by Gene. Bio, and, very recently, by NIH. Web server: http: //www. expasy. ch/sprot-top. html

SWISS-PROT characteristics Almost no redundancy. Cross-references with 60 other databanks. High-quality annotations: Systematic control by a team of annotators. Help from a set of > 200 volunteer experts. Embedded in Expasy, a www proteomics server (http: //www. expasy. org).
Annotations Protein function. Post-translational modifications. Structural or functional domains. Secondary and quaternary structures. Similarities with other proteins. Conflicts between positions for CDS. Disease-related mutations
Associated databanks Tr. EMBL, built using only annotated CDS from the EMBL data library. ENZYME, for the international enzyme nomenclature. PROSITE, for biologically significant sites, patterns and profiles. SWISS-2 DPAGE, for two-dimensional polyacrylamide gel electrophoresis maps.
PIR (The Protein Information Resource) was created by Margaret Dayhoff in 1965. Aims: To provide exhaustive and non-redundant protein sequence data. To give a classification using taxonomic and similarity data: entries grouped in super-families, families and subfamilies.

Data maintenance Three organisms collect and organize the data introduced in PIR: The National Biomedical Research Foundation (NBRF) in the United States. The Martinsried Institute for Protein Sequence (MIPS) in Germany. The Japan International Protein Sequence Information Database (JIPID) in Japan.

Results The exhaustivity is not better than what is obtained with SWISS-PROT+Tr. EMBL. Still contains redundancy. Less comprehensive annotation. Low number of cross-references. PIR has recently joined forces with EBI and SIB to establish the Uni. Prot (United Protein Databases), the central resource of protein sequence and function.

Specialized databanks A lot of specialized databanks have been developed, which are devoted to: Complete genomes. Families of homologous genes. Non-sequence data. These systems are under the responsibility of curators: Data quality and homogeneity control.

Complete genomes There is a large number of databanks devoted to specific organisms. These banks are associated to sequencing or mapping projects. For some model organisms there are often several concurrent systems.

Examples Organism Available databanks Bacillus subtilis NRSub (Non-Redundant B. subtilis) Subti. List Escherichia coli Colibri Eco. Gene (E. coli Gene Database) ECDC (E. coli Database Collection) Various prokaryotes CMR (Comprehensive Microbial Resource) EMGLib (Enhanced Microbial Genomes Library) Micado (Microbial Advanced Database Organization) Saccharomyces cerevisiae MYGD (MIPS Yeast Genome Database) SGD (Saccharomyces Genome Database) YPD (Yeast Proteome Database) Drosophila melanogaster Fly. Base Plasmodium falciparum Plasmo. DB (P. falciparum Database) Caenorhabditis elegans Worm. Base Worm. PD (Worm Protein Database) Arabidopsis thaliana TAIR (The Arabidopsis Information Resource)
Gene family databanks Built with automated procedures: Similarity search between sets of proteins (BLASTP, FASTP, Smith-Waterman). Clustering into homologous families using similarity criteria. Include various data: Protein (and sometimes nucleotide) sequences. Multiple sequence alignments and trees. Taxonomy.
Prot. Fam Developed at MIPS. Built with PIR sequences. Includes four levels of classification: Superfamilies (based on function and similarity criteria). Families (50% similarity). Subfamilies (80% similarity). Entries (≥ 95% similarity).

Prot. FAm characteristics Allows to visualize alignments and dendrograms for the families. Integrates Pfam domains. Allows users to classify their own protein sequences. Web server: http: //mips. gsf. de
Proto. Map Initially developed at the Hebrew University of Jerusalem ; now hosted at Cornell University. Built with SWISS-PROT & Tr. EMBL sequences. Combines 3 sequence similarity measures (BLASTP, FASTA and Smith-Waterman).
Proto. Map characteristics Alignments and trees are visualized with Java applets. Users can submit sequences and classify them. Web server: http: //protomap. cornell. edu/index. html
Specialized systems HOVERGEN (Homologous Vertebrate Genes Database) : HOBACGEN (Homologous Bacterial Genes Database) for prokaryotes and yeast: Based on Gen. Bank CDS. Based on SWISS-PROT/Tr. EMBL. HOBACGEN-CG for completely sequenced genomes: Based on SWISS-PROT/Tr. EMBL.
Other specialized systems COG (Clusters of Orthologous Groups), also for complete genomes: Nu. Re. Base (Nuclear Receptors Database) for mammalian nuclear receptors: Based on Gen. Bank CDS. Based on EMBL CDS. RTKdb (Tyrosine Kinase Receptors): Based on EMBL CDS.
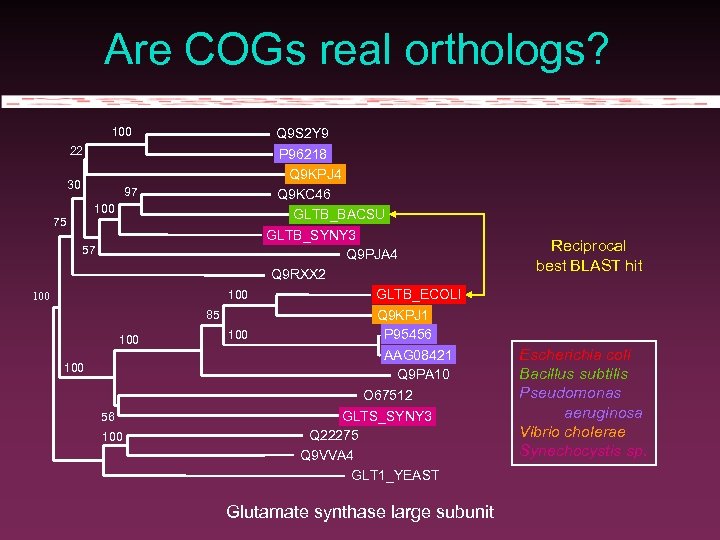
Are COGs real orthologs? 100 Q 9 S 2 Y 9 P 96218 Q 9 KPJ 4 Q 9 KC 46 GLTB_BACSU 22 30 97 100 75 57 100 85 100 56 100 GLTB_SYNY 3 Q 9 PJA 4 Q 9 RXX 2 GLTB_ECOLI Q 9 KPJ 1 P 95456 AAG 08421 Q 9 PA 10 O 67512 GLTS_SYNY 3 Q 22275 Q 9 VVA 4 GLT 1_YEAST Glutamate synthase large subunit Reciprocal best BLAST hit Escherichia coli Bacillus subtilis Pseudomonas aeruginosa Vibrio cholerae Synechocystis sp.

Beyond protein families Prot. Fam, Hovergen, Hobacgen, COGs gather protein sequences homologous on their whole length Patterns, profiles, domains, … are covered in Terry Attwood’s lecture.
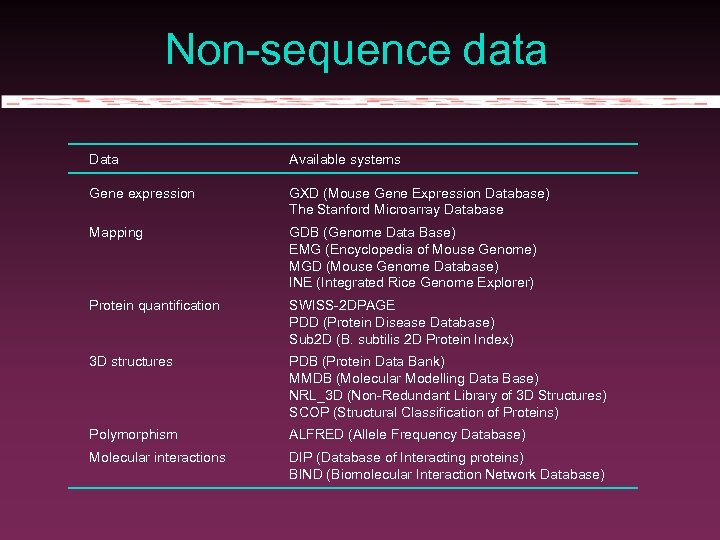
Non-sequence data Data Available systems Gene expression GXD (Mouse Gene Expression Database) The Stanford Microarray Database Mapping GDB (Genome Data Base) EMG (Encyclopedia of Mouse Genome) MGD (Mouse Genome Database) INE (Integrated Rice Genome Explorer) Protein quantification SWISS-2 DPAGE PDD (Protein Disease Database) Sub 2 D (B. subtilis 2 D Protein Index) 3 D structures PDB (Protein Data Bank) MMDB (Molecular Modelling Data Base) NRL_3 D (Non-Redundant Library of 3 D Structures) SCOP (Structural Classification of Proteins) Polymorphism ALFRED (Allele Frequency Database) Molecular interactions DIP (Database of Interacting proteins) BIND (Biomolecular Interaction Network Database)
Sequence Data retrieval Made mainly through Internet access: With client software (e. g. , Entrez, Hobac. Fetch). By remote connections to servers providing online access to the banks (INFOBIOGEN). Using World-Wide Web servers and browsers
Advantages and limitations Users do not have to cope with the usual databases problems: Storing of large amounts of data. Daily updates. Software upgrades. Simplicity of use. Net access is sometimes very slow at peak hours: consider using other servers besides NCBI

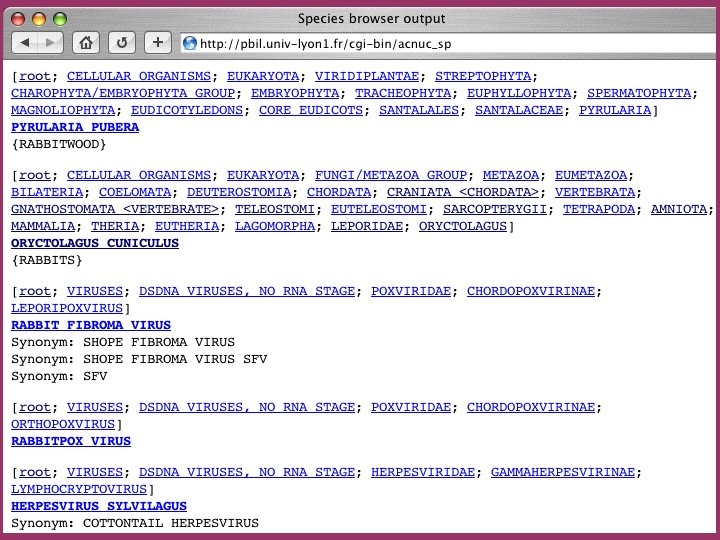
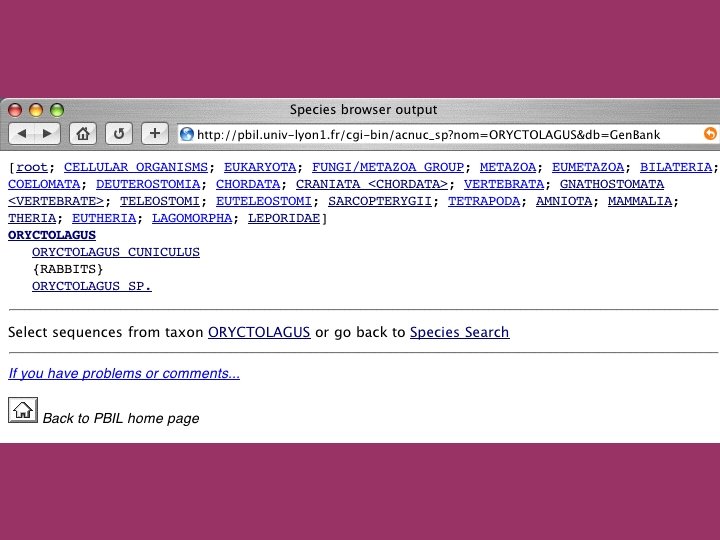
The ACNUC retrieval system Direct access to functional regions described in feature tables (CDS, t. RNA, r. RNA). Selection of entries using various criteria: Sequence names and accession numbers. Bibliographic criteria. Keywords. Taxonomy. Organelle. Developed at Lyon University

ACNUC : possible accesses Graphical interface distributed along with the databases themselves. http: //pbil. univ-lyon 1. fr/databases/acnuc. html Web access at Pôle Bio-Informatique Lyonnais (PBIL): http: //pbil. univ-lyon 1. fr/search/query. html

ACNUC characteristics Allows to query any bank in PIR, SWISSPROT, EMBL, or Gen. Bank formats. Keywords and species browsing. Complex queries. Links with sequence analysis programs on the Web server (alignment, codon usage).
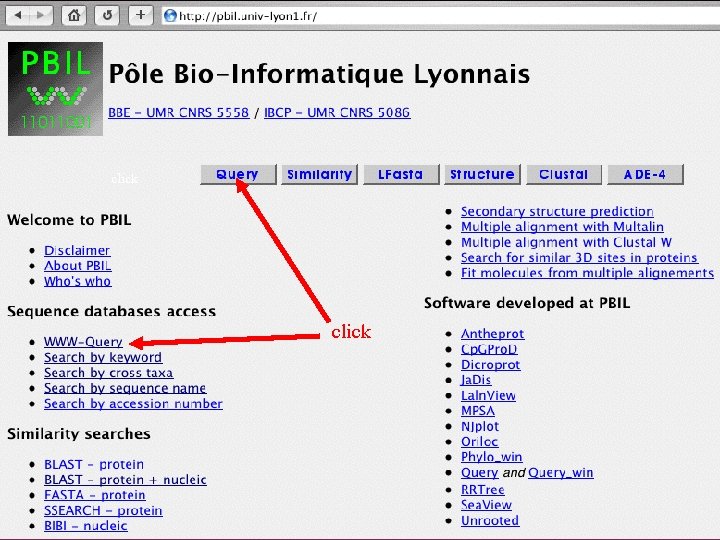
click
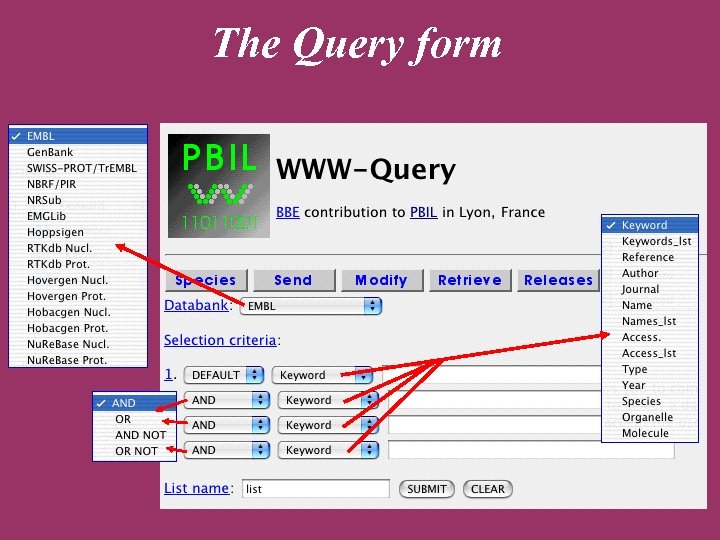
The Query form
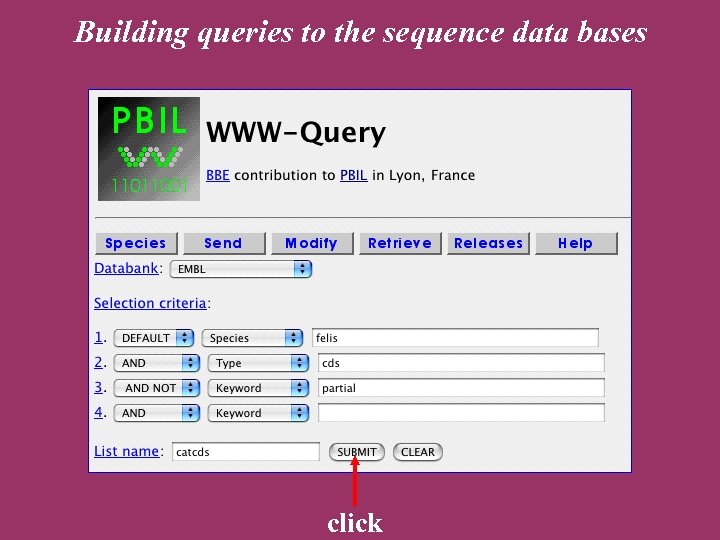
Building queries to the sequence data bases click
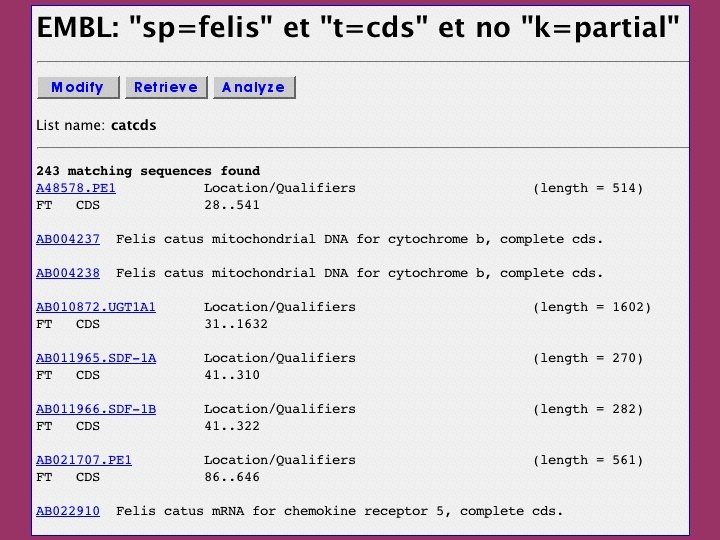

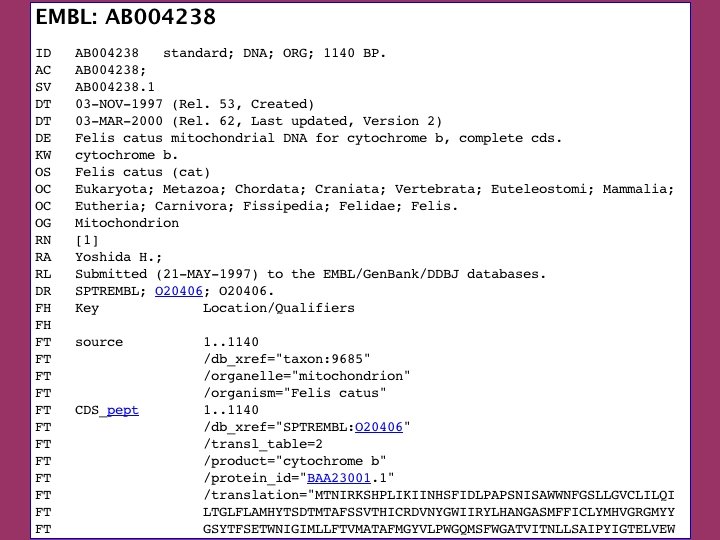
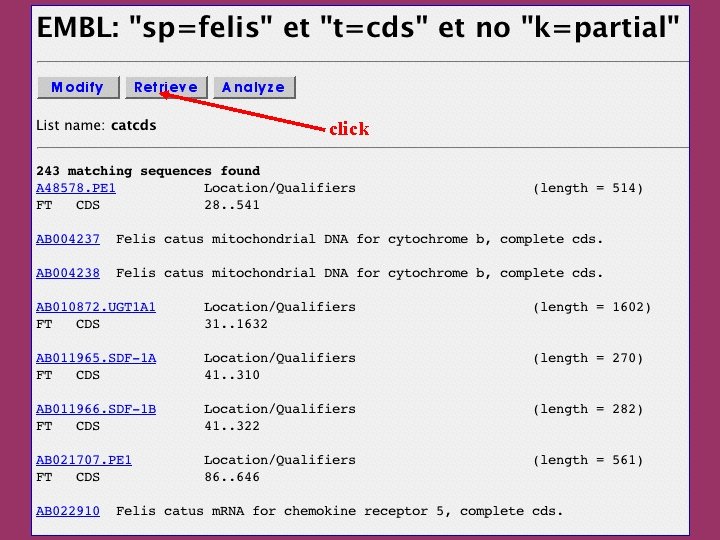
click
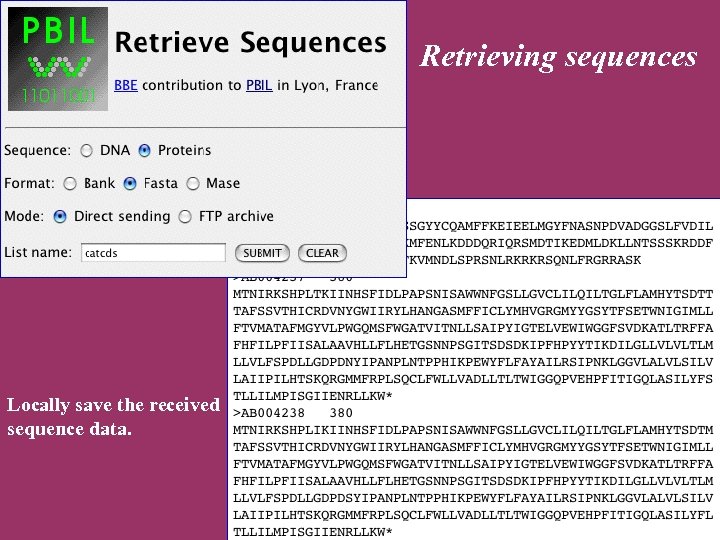
Retrieving sequences Locally save the received sequence data.
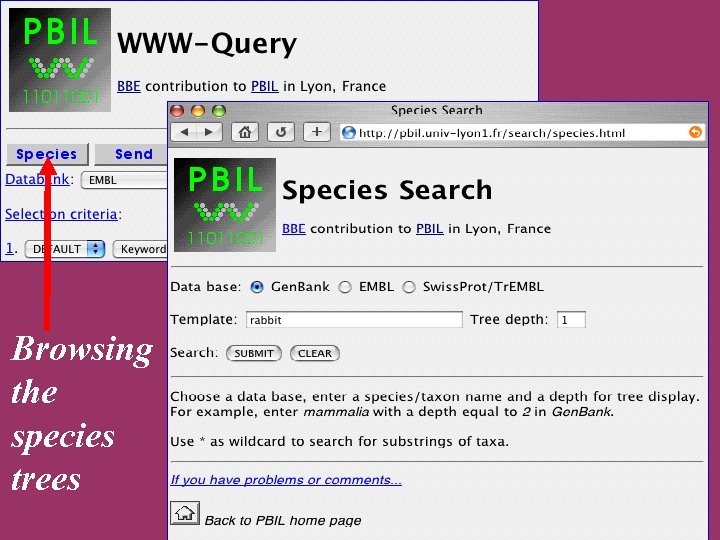
Browsing the species trees
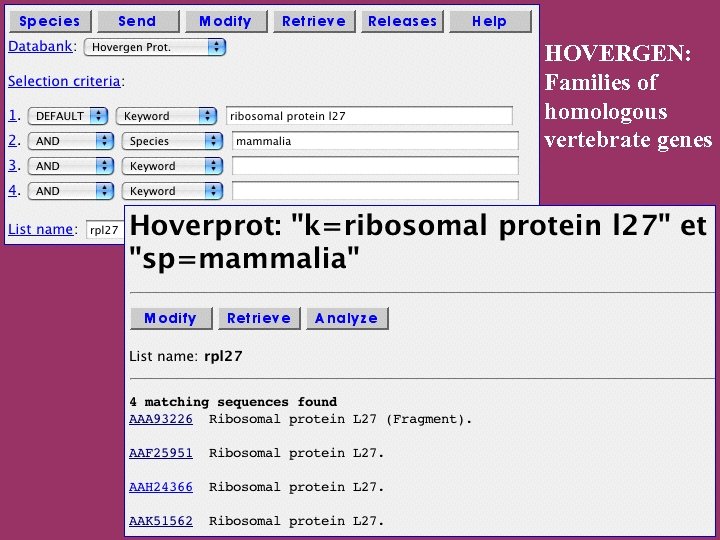
HOVERGEN: Families of homologous vertebrate genes
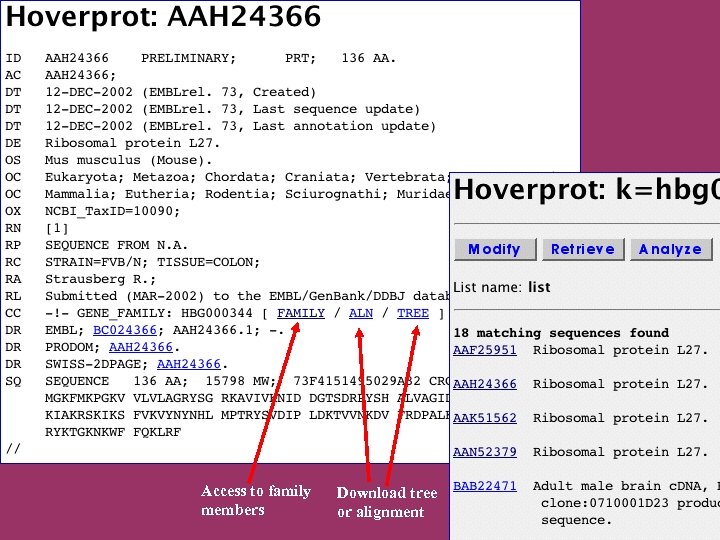
Access to family members Download tree or alignment
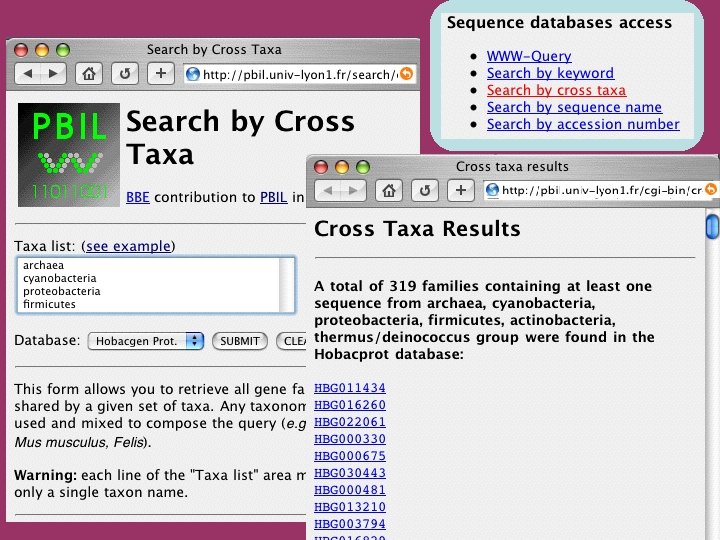

SRS Public version developed at EMBL by Etzold and Argos (1993). Presently available on the different Web servers belonging to EMBnet: EBI (England). INFOBIOGEN (France). DKFZ (Germany). …
Characteristics Database index built with the use of ODD (Object Design and Definition). More than 250 databanks have been indexed and are accessible through 35 SRS servers. Allows queries to operate simultaneously on different banks.
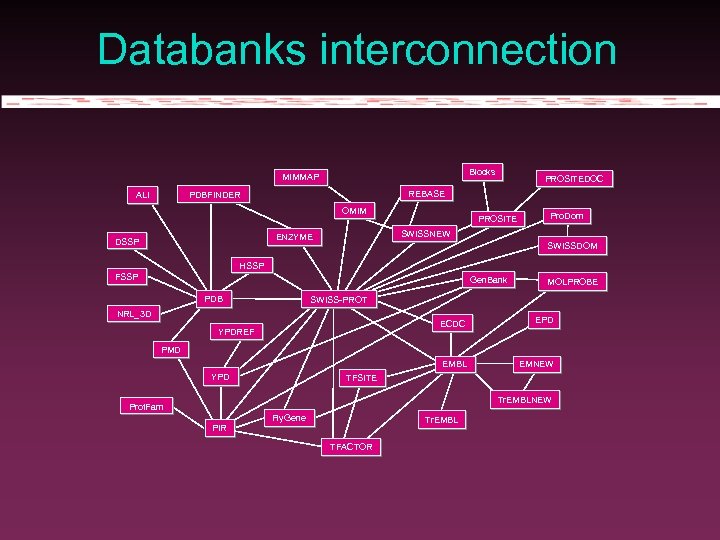
Databanks interconnection Blocks MIMMAP REBASE PDBFINDER ALI OMIM PROSITE Pro. Dom SWISSNEW ENZYME DSSP PROSITEDOC SWISSDOM HSSP FSSP Gen. Bank PDB MOLPROBE SWISS-PROT NRL_3 D ECDC EPD EMBL YPDREF EMNEW PMD YPD TFSITE Tr. EMBLNEW Prot. Fam Fly. Gene Tr. EMBL PIR TFACTOR

Entrez Developed by Schuler et al. (1996) at NCBI. Allows to query several US-made databases: Gen. Bank, Gen. Pept, NR, MMDB, MEDLINE. Access through client software (Unix, Mac or Windows) or Web server: http: //www. ncbi. nlm. nih. gov
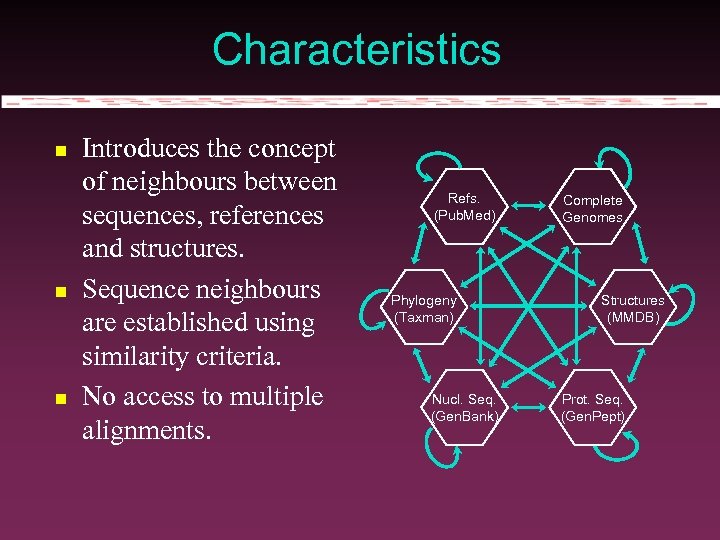
Characteristics Introduces the concept of neighbours between sequences, references and structures. Sequence neighbours are established using similarity criteria. No access to multiple alignments. Refs. (Pub. Med) Phylogeny (Taxman) Nucl. Seq. (Gen. Bank) Complete Genomes Structures (MMDB) Prot. Seq. (Gen. Pept)

NAR 2003 database issue http: //nar. oupjournals. org/content/vol 31/issue 1/
29abbe9a5219a7b08e8f8b93571dc640.ppt