c9917b2efd17b48e8b7a6f389f254131.ppt
- Количество слайдов: 76
SEQUENCE ANALYZES Dilvan Moreira (based on Prof. André Carvalho presentation)

Reading Introduction to Computational Genomics: A Case Studies Approach Chapter 1
Introduction 3 Cells Molecular Biology Probabilistic Sequence Models Multinomiais Models Markov Models Genome Annotation Data Base André de Carvalho - ICMC/USP 3/17/2018
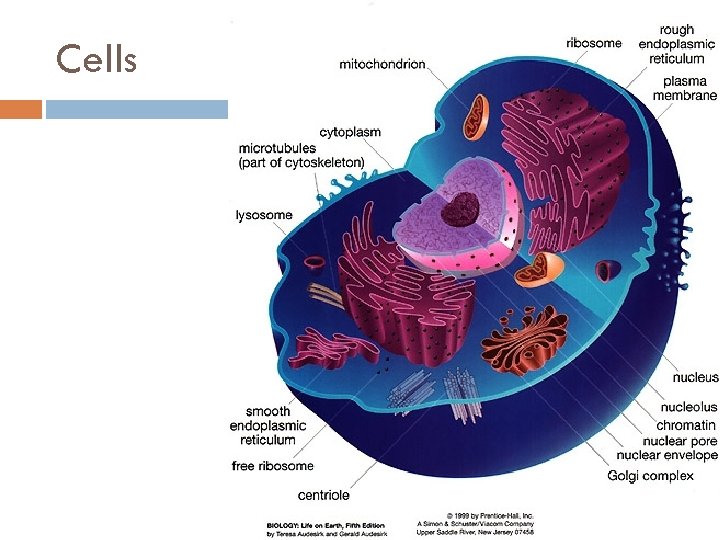
Cells
Cells 5 Cell Basic unity of all living being Compartment involved by membrane, filled with aqueous solution May have organelles with specific functions Mitochondria: energy generation Golgi Complex: accumulation of secretions Among others André de Carvalho - ICMC/USP 17/03/2018

Cells 6 Cell Doctrines All living beings are made of cells and its products Cells have structure and function All the cells emerge from pre existing cells One cell can made copies of itself by replication and division André de Carvalho - ICMC/USP 17/03/2018
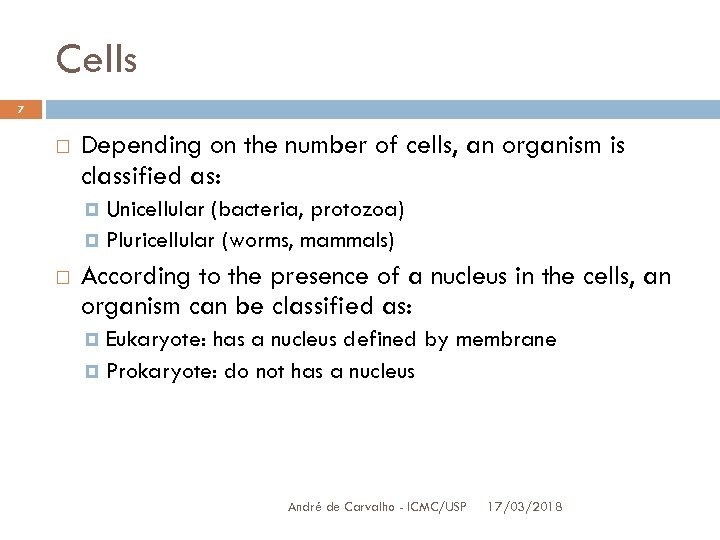
Cells 7 Depending on the number of cells, an organism is classified as: Unicellular (bacteria, protozoa) Pluricellular (worms, mammals) According to the presence of a nucleus in the cells, an organism can be classified as: Eukaryote: has a nucleus defined by membrane Prokaryote: do not has a nucleus André de Carvalho - ICMC/USP 17/03/2018

Cells 8 The fact that an organism is a prokaryote does not mean it is unicellular The majority lives as a unicellular organism Although some species group in a “bunch”, chains or other organization forms of multicellular structures Many unicellular organisms are eukaryotes André de Carvalho - ICMC/USP 3/17/2018
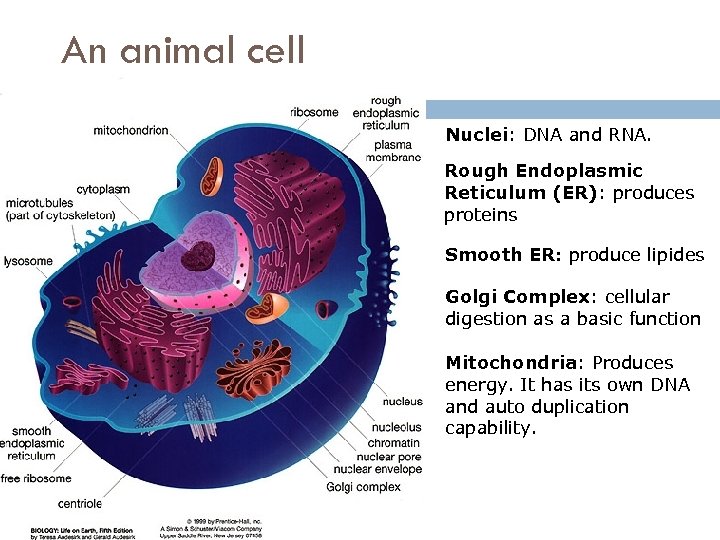
An animal cell Nuclei: DNA and RNA. Rough Endoplasmic Reticulum (ER): produces proteins Smooth ER: produce lipides Golgi Complex: cellular digestion as a basic function Mitochondria: Produces energy. It has its own DNA and auto duplication capability. 3/17/2018 André de Carvalho - ICMC/USP 9
Cells 10 All the cells of the same organism have the same genes Not all the cells have the same organelles in equal proportions Cells vary in form and function Normally, the form is related to function Specific cell function and form are defined by its expressed genes André de Carvalho - ICMC/USP 3/17/2018
Cells 11 The chemical processes that occur in a cell are basically the same For all the cell types and organisms Even though those cells present different forms and functions The DNA replication in a bacteria is similar to the DNA replication in a mammal It makes scientific advances easier Allowing experiments made with basal living to be used to infer results for other beings André de Carvalho - ICMC/USP 3/17/2018
Cytology X Molecular Biology 12 Cytology Science that studies the cell (fixed) Studies the cellular organization, types, functioning, division mechanism, etc With science advances, it is possible to analyze living cells (in vivo) Molecular level Originated the term: Molecular Biology André de Carvalho - ICMC/USP 3/17/2018
DNA 13 Deoxyribonucleic Acid May have single or double-stranded Double-stranded DNA two strands twisted around each other to form a double helix The long polymer compacts itself in a chromosome The DNA is composed by four different nucleotides (bases) Adenine, Cytosine, Guanine and Thymine (Uracil on RNA) The double-strand is caused by base pairing André de Carvalho - ICMC/USP 3/17/2018

DNA 14 The DNA strands are kept together by links that connect each nucleotide of one strand to its complement in the other strand André de Carvalho - ICMC/USP 3/17/2018

DNA The DNA is always read from the 5‘ end to the 3‘ end in the transcriptional process 5’ ATTTAGGCC 3’ 3’ TAAATCCGG 5’ 17/03/2018 André de Carvalho - ICMC/USP 15
DNA 5’ end In one end, there is the first nucleotide. It has a phosphate group C 5 projecting out. 3’end In the other end, there is the last nucleotide added to the DNA strand. It is the only one that still has the component C 3–OH. 17/03/2018 André de Carvalho - ICMC/USP 16
Molecular Biology 17 The genome is the set of all DNA from a cell (organism) Including the genes Genes carry the necessary information to produce the required proteins of an organism The proteins determine Organism’s appearance How the body metabolizes food or defend itself from infections Sometimes, the organism behavior André de Carvalho - ICMC/USP 3/17/2018
Fraction of yeast genome 18 CCACACCCACACACCACACACCACACCCACACATCCTAACACTACCCTAACACAGCCCTAATCTAACCCTGGCCAACCTGTCTCTCAACTTACCC TCCATTACCCTGCCTCCACTCGTTACCCTGTCCCATTCAACCATACCACTCCGAACCACCATCCCTCTACTACCACTCACCGTTACCCTCCAATTACCCATATCCAACCCAC TGCCACTTACCCTACCATCCACCATGACCTACTCACCATACTGTTCTTCTACCCACCATATTGAAACGCTAACAAATGATCGTAAATAACACGTGCTTACCCTACCACT TTATACCACCACCACATGCCATACTCACCCTCACTTGTATACTGATTTTACGCACACGGATGCTACAGTATATACCATCTCAAACTTACCCTACTCTCAGATTCCACTTCACTCCATGGC CCATCTCTCACTGAATCAGTACCAAATGCACTCACATCATTATGCACGGCACTTGCCTCAGCGGTCTATACCCTGTGCCATTTACCCATAACGCCCATCATTATCCACATTTTGATATCTATAT CTCATTCGGCGGTCCCAAATATTGTATAACTGCCCTTAATACGTTATACCACTTTTGCACCATATACTTACCACTCCATTTATATACACTTATGTCAATATTACAGAAAAATCCCCACAA AAATCACCTAAACATAAAAATATTCTACTTTTCAACAATAATACATAAACATATTGGCTTGTGGTAGCAACACTATCATGGTATCACTAACGTAAAAGTTCCTCAATATTGCAATTTGCTTGAA CGGATGCTATTTCAGAATATTTCGTACTTACACAGGCCATACATTAGAATAATATGTCACATCACTGTCGTAACACTCTTTATTCACCGAGCAATAATACGGTAGTGGCTCAAACTCATGCGGG TGCTATGATACAATTATATCTTATTTCCATTCCCATATGCTAACCGCAATATCCTAAAAGCATAACTGATGCATCTTTAATCTTGTATGTGACACTACTCATACGAAGGGACTATATCTAGTCA AGACGATACTGTGATAGGTACGTTATTTAATAGGATCTATAACGAAATGTCAAATAATTTTACGGTAATATAACTTATCAGCGGCGTATACTAAAACGGACGTTACGATATTGTCTCACTTCAT CTTACCACCCTCTATCTTATTGCTGATAGAACACTAACCCCTCAGCTTTATTTCTAGTTACACAAAAAACTATGCCAACCCAGAAATCTTGATATTTTACGTGTCAAAAAATGAGGGT CTCTAAATGAGAGTTTGGTACCATGACTTGTAACTCGCACTGCCCTGATCTGCAATCTTGTTCTTAGAAGTGACGCATATTCTATACGGCCCGACGCGCCAAAAAATGAAAAACGAAGC AGCGACTCATTTTTAAGGACAAAGGTTGCGAAGCCGCACATTTCCAATTTCATTGTTGTTTATTGGACATACACTGTTAGCTTTATTACCGTCCACGTTTTTTCTACAATAGTGTAGAAG TTTCTTATGTTCATCGTATTCATAAAATGCTTCACGAACACCGTCATTGATCAAATAGGTCTATAATATACATTTATATAATCTACGGTATTTATATCATCAAAAGTAGT TTTTTTATTTTGTTCGTTAATTTTCAATTTCTATGGAAACCCGTTCGTAAAATTGGCGTTTGTCTCTAGTTTGCGATAGTGTAGATACCGTCCTTGGATAGAGCACTGGAGATGGCTGG CTTTAATCTGCTGGAGTACCATGGAACACCGGTGATCATTCTGGTCACTTGGTCTGGAGCAATACCGGTCAACATGGTGGTGAAGTCACCGTAGTTGAAAACGGCTTCAGCAACTTCGACTGGG TAGGTTTCAGTTGGGCGGCTTGGAACATGTAGTATTGGGCTAAGTGAGCTCTGATATCAGAGACGTAGACACCCAATTCCACCAAGTTGACTCTTTCGTCAGATTGAGCTAGAGTGGTGG TTGCAGAAGCAGTAGCAGCGATGGCAGCGACACCAGCGGCGATTGAAGTTAATTTGACCATTGTATTTGTTTGTTAGTGCTGATATAAGCTTAACAGGAAAGAATAAAGACATA TTCTCAAAGGCATATAGTTGAAGCAGCTCTATTTATACCCATTCCCTCATGGGTTGTTGCTATTTAAACGATCGCTGACTGGCACCAGTTCCTCATCAAATATTCTCTATATCTCATCTTTCAC ACAATCTCATTATCTCTATGGAGATGCTCTTGTTTCTGAACGAATCATAAATCTTTCATAGGTTTCGTATGTGGAGTACTGTTTTATGGCGCTTATGTGTATTCGTATGCGCAGAATGTGGGAA TGCCAATTATAGGGGTGCCGAGGTGCCTTATAAAACCCTTTTCTGTGCCTGTGACATTTCCTTTTTCGGTCAAAAAGAATATCCGAATTTTAGATTTGGACCCTCGTACAGAAGCTTATTGTCT AAGCCTGAATTCAGTCTGCTTTAAACGGCTTCCGCGGAGGAAATATTTCCATCTCTTGAATTCGTACAACATTAAACGTGTGTTGGGAGTCGTATACTGTTAGGGTCTGTAAACTTGTGAACTC TCGGCAAATGCCTTGGTGCAATTACGTAATTTTAGCCGCTGAGAAGCGGATGGTAATGAGACAAGTTGATATCAAACAGATACATATTTAAAAGAGGGTACCGCTAATTTAGCAGGGCAGTATT ATTGTAGTTTGATATGTACGGCTAACTGAACCTAAGTAGGGATATGAGAGTAAGAACGTTCGGCTACTCTTCTAAGTGGGATTTTTCTTAATCCTTGGATTCTTAAAAGGTTATTAAAGT TCCGCACAAAGAACGCTTGGAAATCGCATTCATCAAAGAACAACTCTTCGTTTTCCAAACAATCTTCCCGAAAAAGTAGCCGTTCATTTCCCTTCCGATTTCATTCCTAGACTGCCAAATTTTT CTTGCTCATTTATAATGATAAGAATTGTATTTGTGTCCCATTCTCGTAGATAAAATTCTTGGATGTTAAAAAATTATTATTTTCTTCATAAAGAAGCTTTCAAGATATAAGATACGAAAT AGGGGTTGATAATTGCATGACAGTAGCTTTAGATCAAAAAGGAAAGCATGGAGGGAAACAGTGAAAATTCTCTTGAGAACCAAAGTAAACCTTCATTGAAGAGCTTCCTTAAAAAAT TTAGAATCTCCCATGTCAACGGGTTTCCATACCTCCCCAGCATCATACATCTTTTTTCAAAGAAACTTCAAATGCCTCTTTTATGCAAGGGGCAAAATCCTGAAATGACTTAAACTTAGCAGTT TCGTCTTTTTTCAAAGAGAATGGTTGAAGAAGAATTGTTTTGGACGCTTATTGACAATCTGTTGCATTGATAAAGTACCTACTATCCCAGACTATATTTGTATACAAGTACAAAATTAGGTTTG TTGAAACAACTTTCCGATCATTGGTGCCCGTATCTGATGTTTTTTTAGTAATTTCTTTGTAAATACAGGGAGTTGTTTCGAAAGCTTATGAGAAAAATACATGACAGGTAAAAATATTGG CTCGAAAAAGAGGACAAAAAGAGAAATCATAAATGAGTAAACCCACTTGCTGGACATTATCCAGTAAAGGCTTGGTAGTAACCATAATATTACCCAGGTACGAAACGCTAAGAACCTTGAAAGA CTCATAAAACTTCCAGGTTAAGCTATTTTTGAAAATATTCTGAGGTAAAAGCCATTAAGGTCCAGATAACCAAGGGACAATAAACCTATGCTTTTCTTGTCTTCAATTTCAGTATCTTTCCATT TTGATAATGAGCATGTGATCCGGAAAGCTACTTTATGATGTTTCAAGGCCTGAAGTTTGAATATTTATGTAGTTCAACATCAAATGTGTCTATTTTGTGATGAGGCAACCGTCGACAACCTTAT TATCGAAAAAGAACAACAAGTTCACATGCTTGTTACTCTCTATAACTAGAGAGTACTTTTTTTGGAAGCAAGTAAGAATAAGTCAATTTCTACTTACCTCATTAGGGAAAAATTTAATAGCAGT TGTTATAACGACAAATACAGGCCCTAAAAAATTCACTGTATTCAATGGTCTACGAATCGTCAATCGCTTGCGGTTATGGCACGAAGAACAATGCAATAGCTCTTACAAGCCACTACATGACAAG CAACTCATAATTTAA André de Carvalho - ICMC/USP 17/03/2018
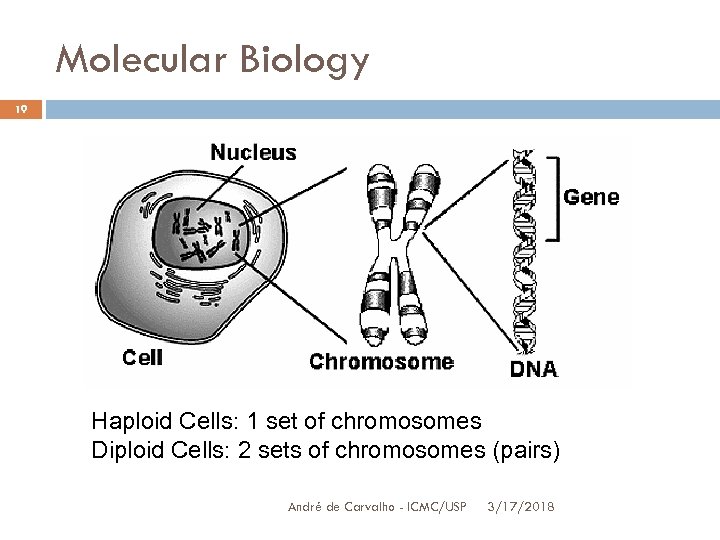
Molecular Biology 19 Haploid Cells: 1 set of chromosomes Diploid Cells: 2 sets of chromosomes (pairs) André de Carvalho - ICMC/USP 3/17/2018
Molecular Biology 20 Genes Subsequences Found of DNA in the chromosome They are the mold of protein or RNA production Between the genes there segments called are noncoding regions André de Carvalho - ICMC/USP 3/17/2018
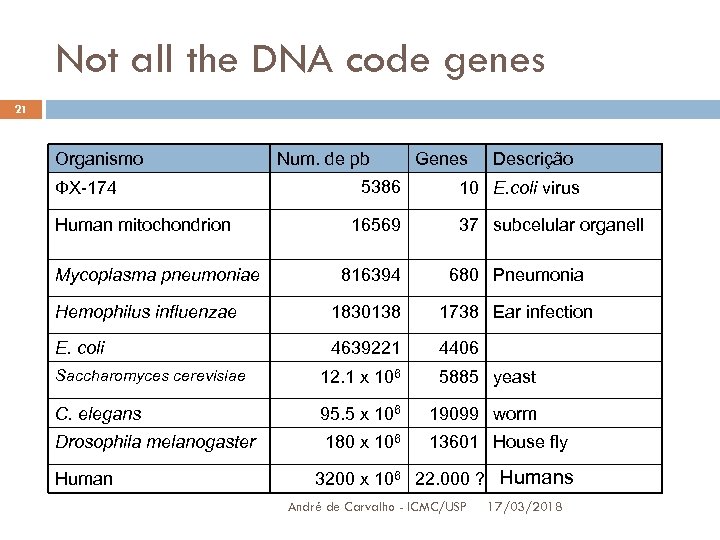
Not all the DNA code genes 21 Organismo ФX-174 Human mitochondrion Mycoplasma pneumoniae Num. de pb 5386 16569 Genes Descrição 10 E. coli virus 37 subcelular organell 816394 680 Pneumonia Hemophilus influenzae 1830138 1738 Ear infection E. coli 4639221 4406 Saccharomyces cerevisiae 12. 1 x 106 5885 yeast C. elegans 95. 5 x 106 19099 worm Drosophila melanogaster 180 x 106 13601 House fly Human 3200 x 106 22. 000 ? Humans André de Carvalho - ICMC/USP 17/03/2018
Non-coding DNA 22 It is not part of protein/RNA synthesis It was considered genetic “junk” It binds to a DNA strand One of its functions: blocking the transcriptional process The bound gene is not read It avoids the expression of the associated protein Inhibition of genes may prevent tumor cells growth Researchers were able to bind genes not related to tumor growth André de Carvalho - ICMC/USP 3/17/2018
Molecular Biology 23 Scientists identified the gene related to breast cancer (SATB 1) Paper published on Nature (March, 2008) Healthy organism: organizer of other genes Cancerous organism: growth of tumors, controlling around 1000 other genes Gang leader, gang, mob Active role on formation of other cancer focus (metastasis) Most common cause of death in ill patients André de Carvalho - ICMC/USP 3/17/2018
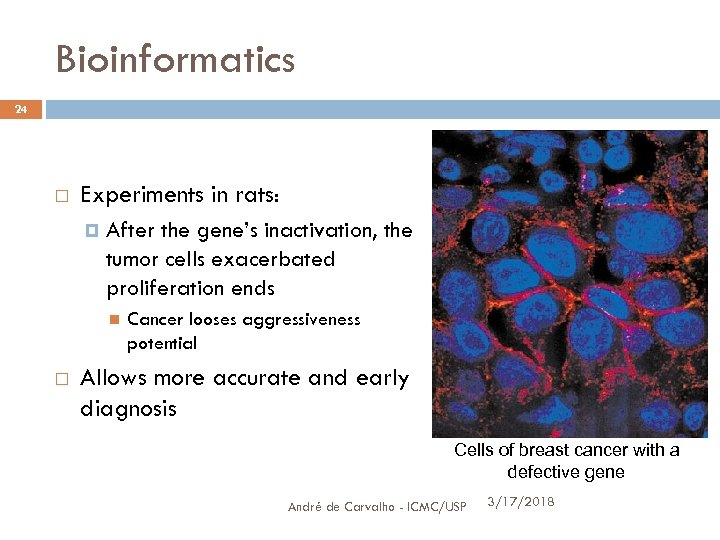
Bioinformatics 24 Experiments in rats: After the gene’s inactivation, the tumor cells exacerbated proliferation ends Cancer looses aggressiveness potential Allows more accurate and early diagnosis Cells of breast cancer with a defective gene André de Carvalho - ICMC/USP 3/17/2018
Molecular Biology 25 Proteins Define cells structure, function and regulatory mechanism of Example of regulatory mechanisms: control of cellular cycle, genic transcription Linear sequences 20 different amino acid combinations Three consecutive nucleotide (codon) form an amino acid André de Carvalho - ICMC/USP 3/17/2018
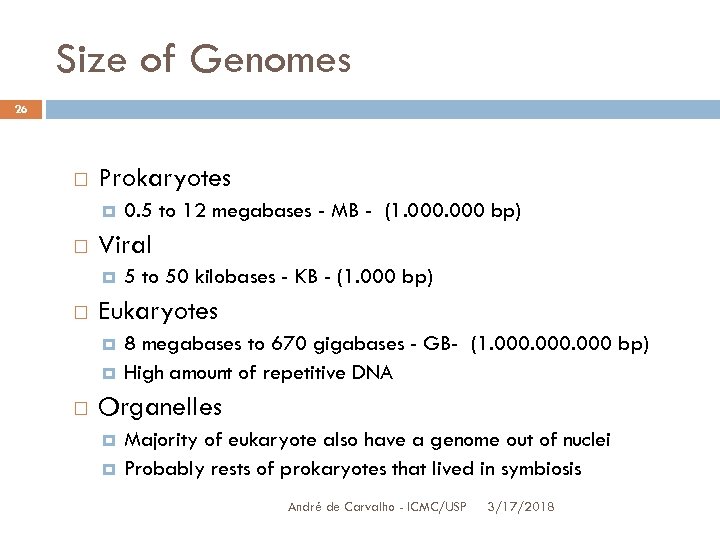
Size of Genomes 26 Prokaryotes Viral 5 to 50 kilobases - KB - (1. 000 bp) Eukaryotes 0. 5 to 12 megabases - MB - (1. 000 bp) 8 megabases to 670 gigabases - GB- (1. 000 bp) High amount of repetitive DNA Organelles Majority of eukaryote also have a genome out of nuclei Probably rests of prokaryotes that lived in symbiosis André de Carvalho - ICMC/USP 3/17/2018
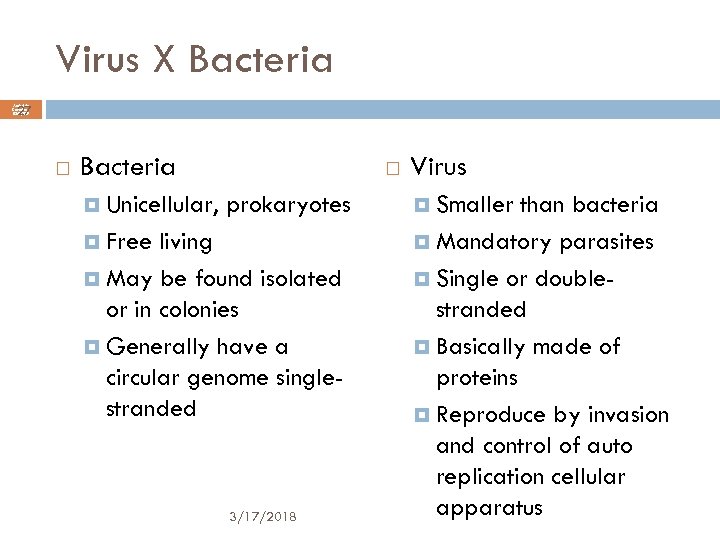
Virus X Bacteria 27 André de Carvalho ICMC/USP Bacteria Unicellular, prokaryotes Free living May be found isolated or in colonies Generally have a circular genome singlestranded 3/17/2018 Virus Smaller than bacteria Mandatory parasites Single or doublestranded Basically made of proteins Reproduce by invasion and control of auto replication cellular apparatus
Probabilistic Models of Sequences 28 The majority of computational genomic studies uses statistical methods Ex. : find structures of interest in sequences of millions bp The majority of the sequence does not have relevant information Need to obtain probabilistic models of DNA sequences André de Carvalho - ICMC/USP 3/17/2018
Probabilistic Models of Sequences 29 Abstraction of the 3 D molecule to a symbol sequence (linear) Alphabet {A, C, T, G} Allows the use of powerful mathematic tools Lack of care with information of the tridimensional structure André de Carvalho - ICMC/USP 3/17/2018
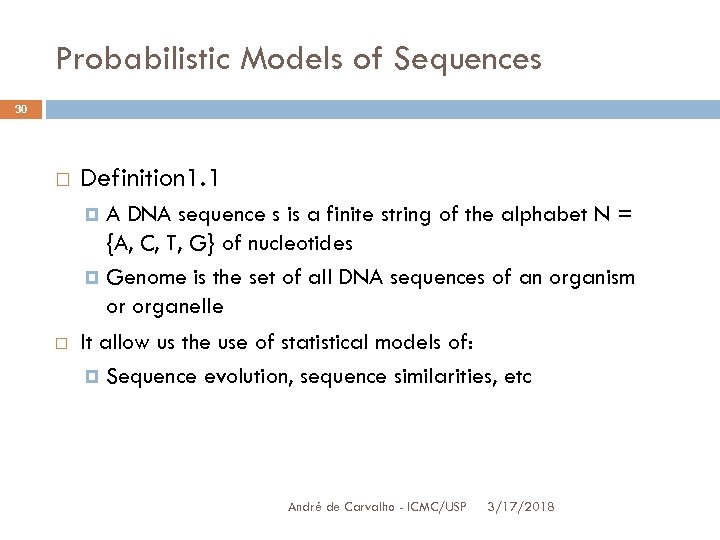
Probabilistic Models of Sequences 30 Definition 1. 1 A DNA sequence s is a finite string of the alphabet N = {A, C, T, G} of nucleotides Genome is the set of all DNA sequences of an organism or organelle It allow us the use of statistical models of: Sequence evolution, sequence similarities, etc André de Carvalho - ICMC/USP 3/17/2018
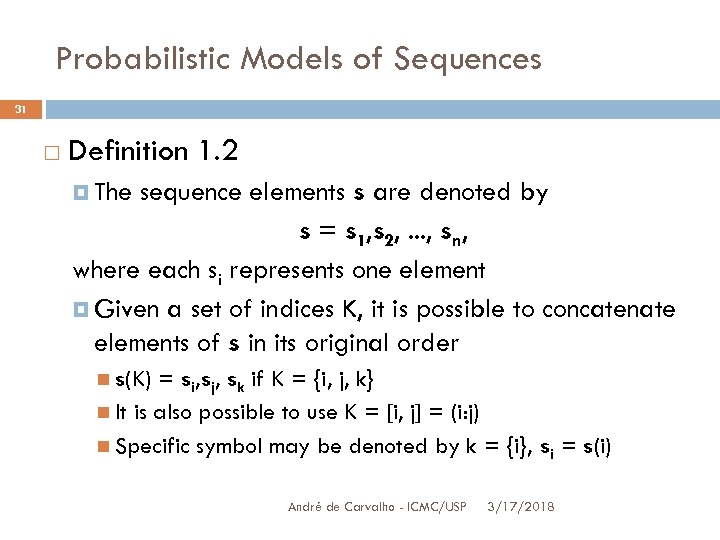
Probabilistic Models of Sequences 31 Definition 1. 2 The sequence elements s are denoted by s = s 1, s 2, . . . , sn, where each si represents one element Given a set of indices K, it is possible to concatenate elements of s in its original order s(K) = si, sj, sk if K = {i, j, k} It is also possible to use K = [i, j] = (i: j) Specific symbol may be denoted by k = {i}, si = s(i) André de Carvalho - ICMC/USP 3/17/2018

Exercise 32 Given a DNA sequence s = ATATGTCGTGCA, find: s{7} = s(2: 6) = s{2, 5, 9} = André de Carvalho - ICMC/USP 3/17/2018
: Probabilistic Models of Sequences 33 Almost all probabilistic methods of sequence analysis can be grouped into two categories: Multinomial Models Markov Models André de Carvalho - ICMC/USP 3/17/2018
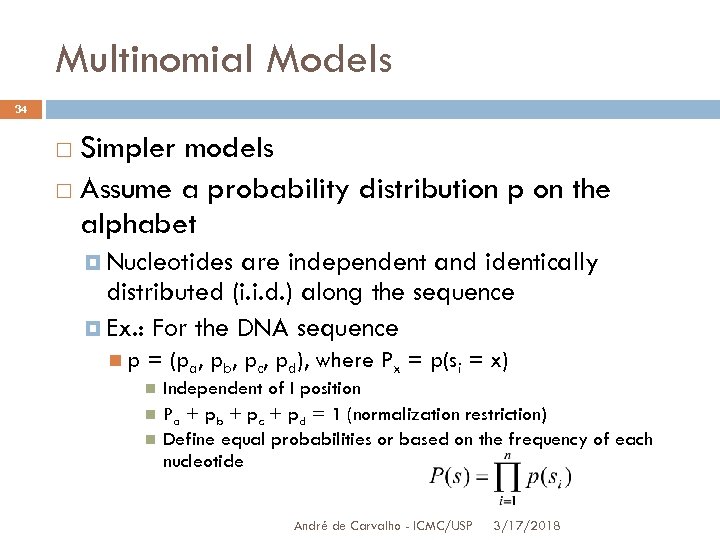
Multinomial Models 34 Simpler models Assume a probability distribution p on the alphabet Nucleotides are independent and identically distributed (i. i. d. ) along the sequence Ex. : For the DNA sequence p = (pa, pb, pc, pd), where Px = p(si = x) Independent of I position Pa + pb + pc + pd = 1 (normalization restriction) Define equal probabilities or based on the frequency of each nucleotide André de Carvalho - ICMC/USP 3/17/2018
Multinomial Models 35 It is not expected that DNA sequences are truly random Model validity can be tested with real sequences Estimate frequency symbols in the sequence regions Test of independence violations checking correlations between neighbor individuals Regions where changes occur and there is interest in the correlations André de Carvalho - ICMC/USP 3/17/2018
Markov Models 36 Provide more complex model of DNA sequences Probability of observing a symbol depends on the previous symbol in the sequence Last symbol – order 1 Last two – order 2 No previous – order 0 (multinomial) Can shape local co-relations between nucleotides André de Carvalho - ICMC/USP 3/17/2018
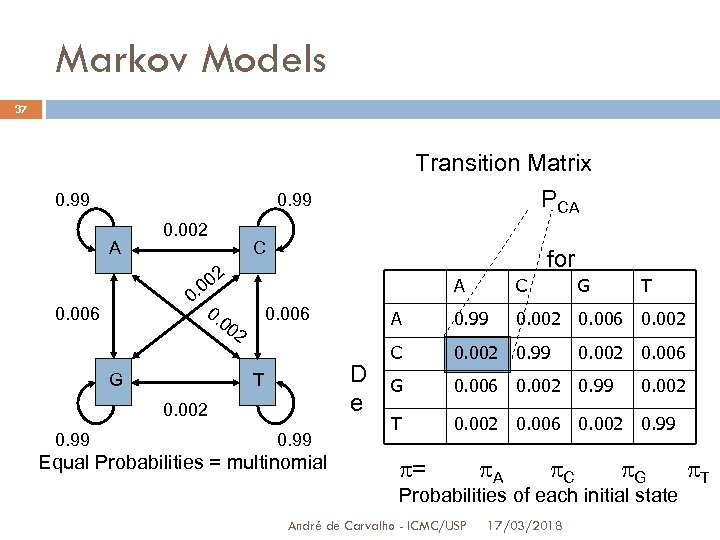
Markov Models 37 0. 99 A 0. 002 C 2 00 0. 0. 00 2 0. 006 G for A D e T 0. 99 Equal Probabilities = multinomial C A 0. 006 0. 002 0. 99 Transition Matrix PCA G T 0. 99 0. 002 0. 006 0. 002 C 0. 002 0. 99 G 0. 006 0. 002 0. 99 T 0. 002 0. 006 0. 002 0. 99 0. 002 0. 006 0. 002 = A C G T Probabilities of each initial state André de Carvalho - ICMC/USP 17/03/2018
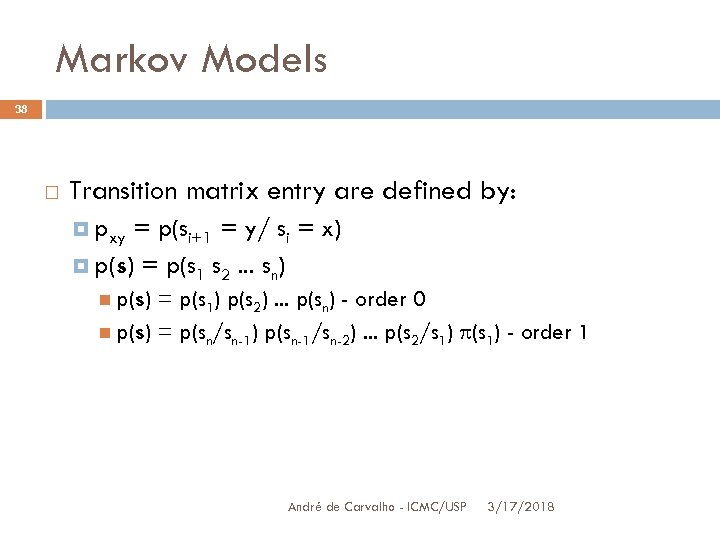
Markov Models 38 Transition matrix entry are defined by: pxy = p(si+1 = y/ si = x) p(s) = p(s 1 s 2. . . sn) p(s) = p(s 1) p(s 2). . . p(sn) - order 0 p(s) = p(sn/sn-1) p(sn-1/sn-2). . . p(s 2/s 1) (s 1) - order 1 André de Carvalho - ICMC/USP 3/17/2018
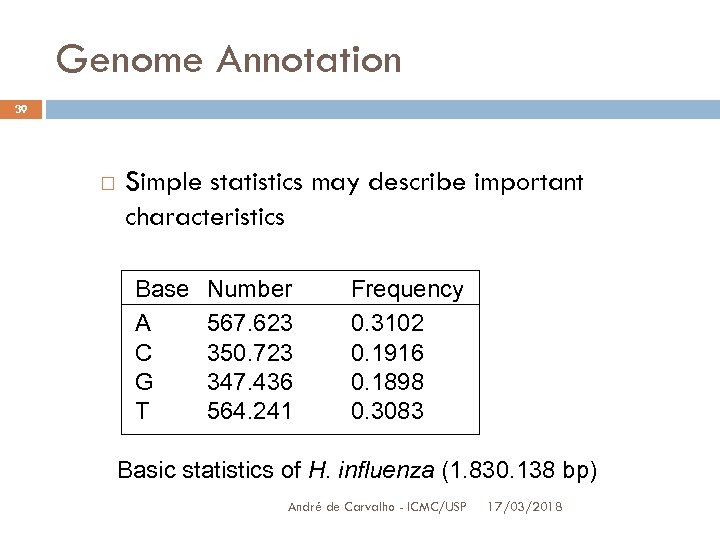
Genome Annotation 39 Simple statistics may describe important characteristics Base A C G T Number 567. 623 350. 723 347. 436 564. 241 Frequency 0. 3102 0. 1916 0. 1898 0. 3083 Basic statistics of H. influenza (1. 830. 138 bp) André de Carvalho - ICMC/USP 17/03/2018
Genome Annotation 40 Base composition Bases frequencies are different in genomes of different organisms Frequencies may vary in different parts Violates multinomial model supposition André de Carvalho - ICMC/USP 3/17/2018
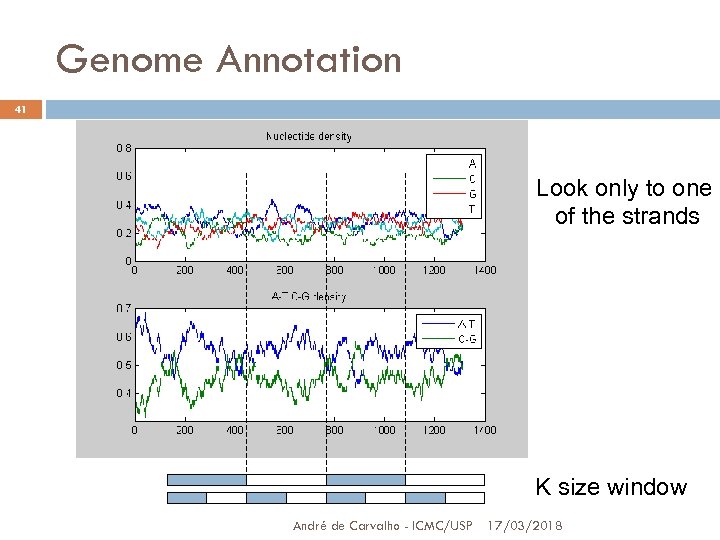
Genome Annotation 41 Look only to one of the strands K size window André de Carvalho - ICMC/USP 17/03/2018
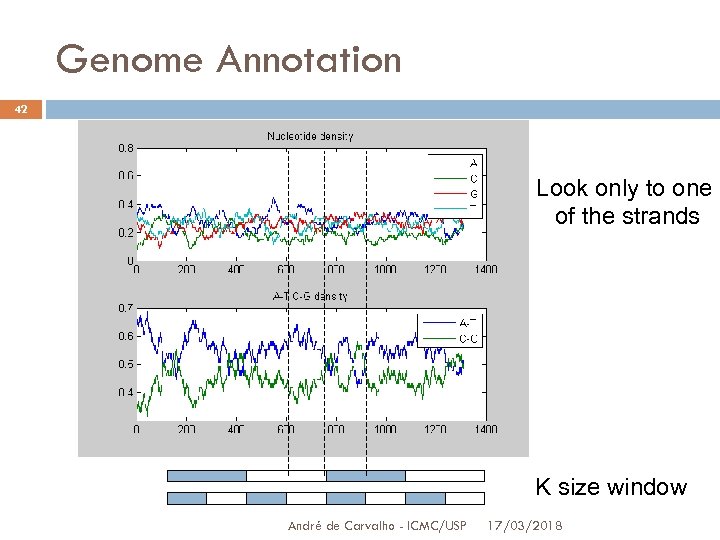
Genome Annotation 42 Look only to one of the strands K size window André de Carvalho - ICMC/USP 17/03/2018
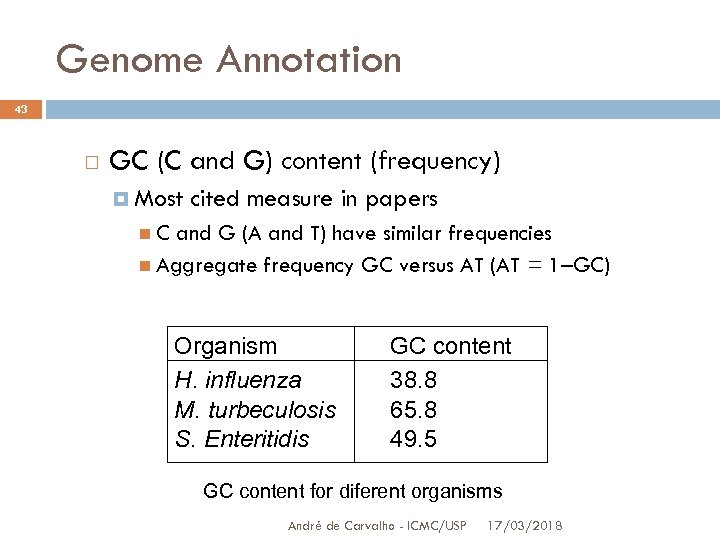
Genome Annotation 43 GC (C and G) content (frequency) Most cited measure in papers C and G (A and T) have similar frequencies Aggregate frequency GC versus AT (AT = 1–GC) Organism H. influenza M. turbeculosis S. Enteritidis GC content 38. 8 65. 8 49. 5 GC content for diferent organisms André de Carvalho - ICMC/USP 17/03/2018
Genome Annotation 44 GC content (frequency) May be used to detect external genetic material in a part of genome Species may acquire sub sequences from other organisms (ex. virus) Horizontal genetic transference André de Carvalho - ICMC/USP 3/17/2018
Genome Annotation 45 Point of change analysis Use this method to detect where the bases distribution (or GC) change These change regions divide the sequence in more uniform parts They may help to identify important biological signs Simpler measure: use threshold Threshold value definition (like the window size) is a statistical problem André de Carvalho - ICMC/USP 3/17/2018
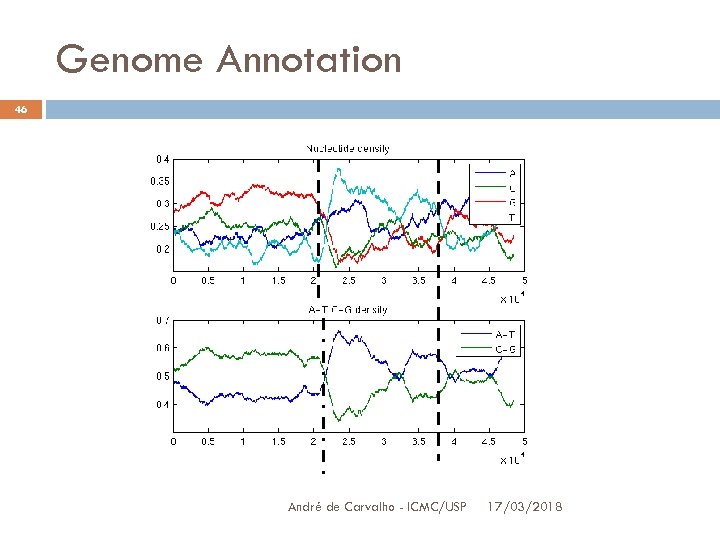
Genome Annotation 46 André de Carvalho - ICMC/USP 17/03/2018
Genome Annotation 47 k-mer frequency is a motif bias Other useful measure is the frequency of sequences size 2 and major Dimers, trimers, k-mers K-mers are not usual: any word that is in the genome with frequency highest or lowest than expected Bias in the position or frequency of these words may reveal important information about its function The number of k-mers is counted by a k-size window covering the sequence André de Carvalho - ICMC/USP 3/17/2018
Genome Annotation 48 k-mer frequency and motif bias It is also possible to plot the frequency of only some interest k-mers Ex. Dimers (dinucleotide) AT and CG There are examples of statistical bias of nucleotides Ex. : Low frequency of CGs in some organisms It is easy to see these bias in “genome signatures” Chaos-Game representation (CGR) André de Carvalho - ICMC/USP 17/03/2018
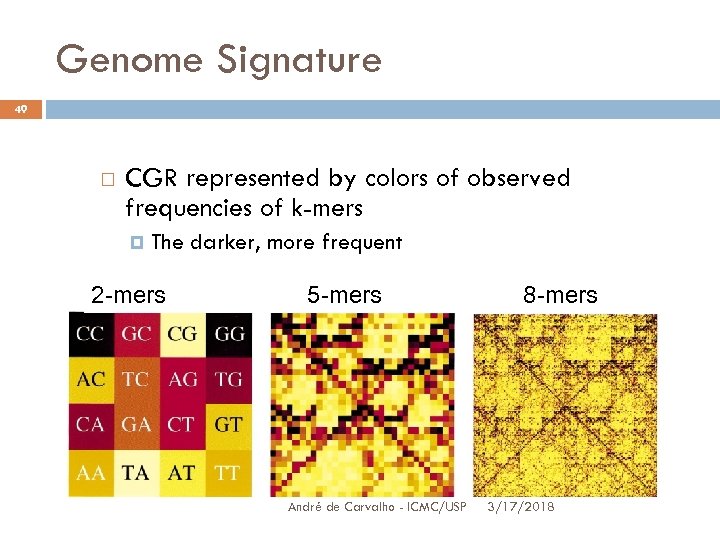
Genome Signature 49 CGR represented by colors of observed frequencies of k-mers The darker, more frequent 2 -mers 5 -mers André de Carvalho - ICMC/USP 8 -mers 3/17/2018
Genome Signature 50 CGR exhibits frequencies of 4 k words or strings The quadrate image is sliced into 4 quadrant q 1, one for each nucleotide A pixel indicating the frequency of all the qq size strings, ending in a given nucleotide, occur in this nucleotide quadrant André de Carvalho - ICMC/USP 3/17/2018
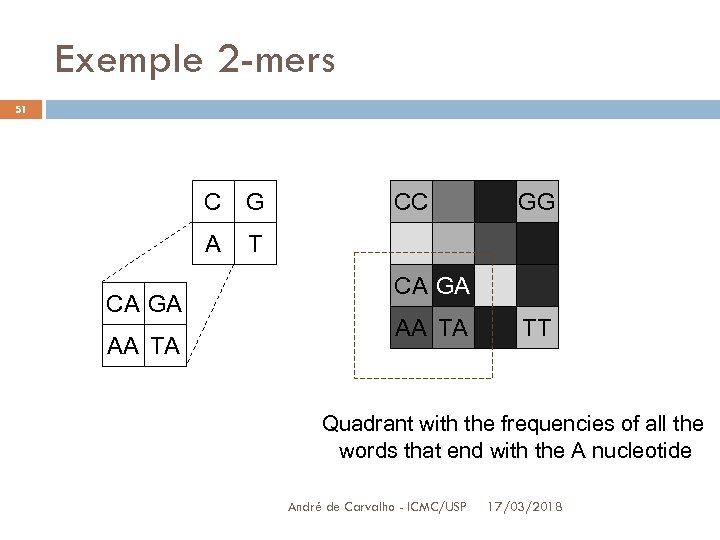
Exemple 2 -mers 51 C A CA GA AA TA G CC GG T CA GA AA TA TT Quadrant with the frequencies of all the words that end with the A nucleotide André de Carvalho - ICMC/USP 17/03/2018
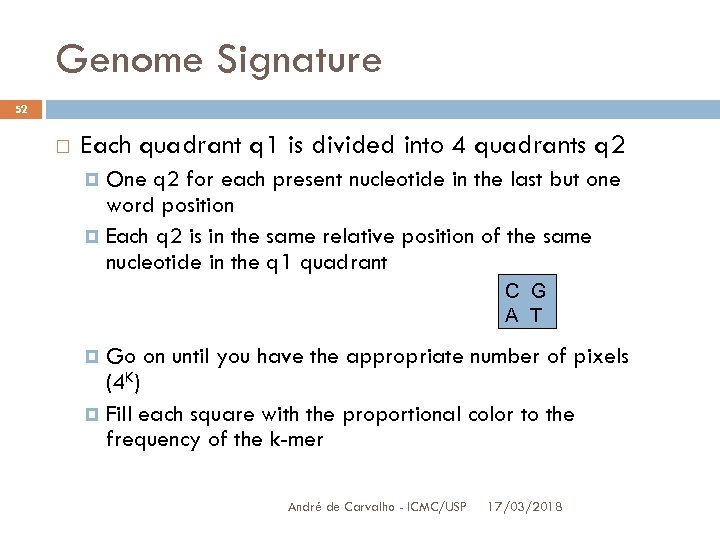
Genome Signature 52 Each quadrant q 1 is divided into 4 quadrants q 2 One q 2 for each present nucleotide in the last but one word position Each q 2 is in the same relative position of the same nucleotide in the q 1 quadrant C G A T Go on until you have the appropriate number of pixels (4 K) Fill each square with the proportional color to the frequency of the k-mer André de Carvalho - ICMC/USP 17/03/2018

Genome Annotation 53 The motifs (k-mers) frequency can bring relevant informations Sequence of frequent nucleotides that may have a biological relevance Simple statistical analysis may consider the nucleotide frequency It can find motifs high or low represented Which helps to decide when the bias is significant or not Non usual motifs may have biological relevance Ex. : motifs may frequently be associated with repetitive elements André de Carvalho - ICMC/USP 3/17/2018
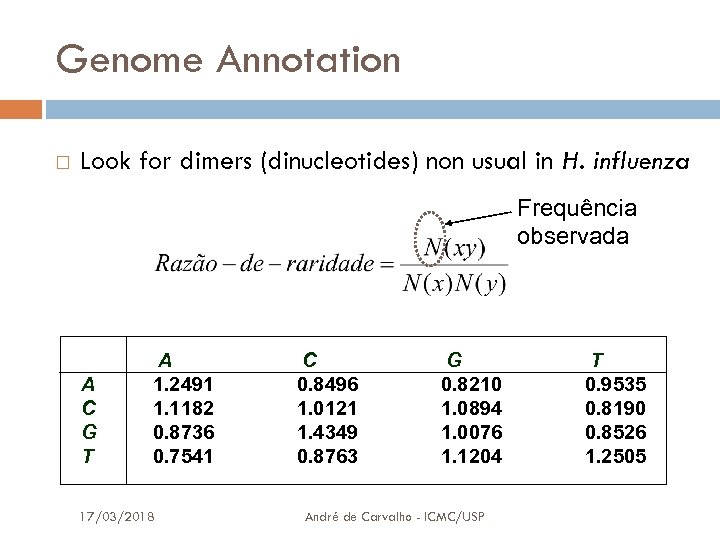
Genome Annotation Look for dimers (dinucleotides) non usual in H. influenza Frequência observada A C G T A 1. 2491 1. 1182 0. 8736 0. 7541 17/03/2018 C 0. 8496 1. 0121 1. 4349 0. 8763 G 0. 8210 1. 0894 1. 0076 1. 1204 André de Carvalho - ICMC/USP T 0. 9535 0. 8190 0. 8526 1. 2505 54
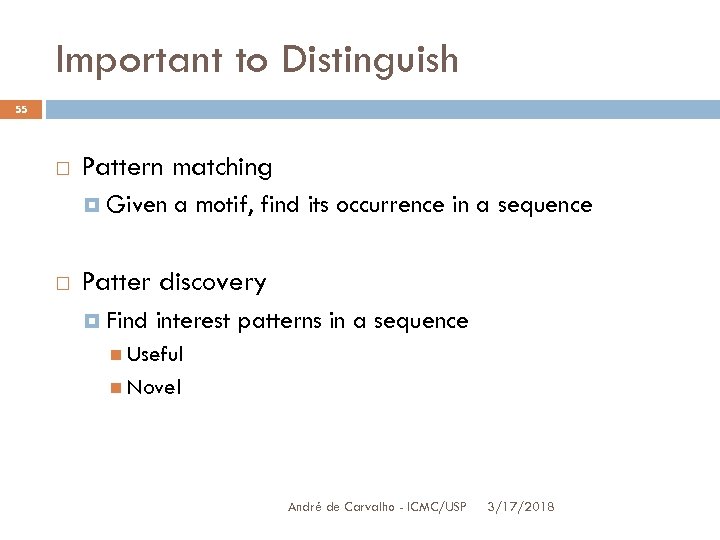
Important to Distinguish 55 Pattern matching Given a motif, find its occurrence in a sequence Patter discovery Find interest patterns in a sequence Useful Novel André de Carvalho - ICMC/USP 3/17/2018
Genome Data. Base 56 The acquired knowledge in this course will be used in real sequences DNA and proteins Stored in DBs available on the internet It is necessary to know how: Access Manipulate Process These data The involved steps are standardized André de Carvalho - ICMC/USP 17/03/2018
Genome Data. Base 57 General DNA, proteins e carbohydrates, 3 -dimension structures , . . . Specialized EST, STS, SNP, RNA, genomes, protein families, pathways, microarray data, . . . ) André de Carvalho - ICMC/USP 17/03/2018
Genome Data. Base 58 All published genome sequence have to be available in a public DB Members of the International Nucleotide Sequence Database Collaboration are the main repositories Consortia made by 3 big DBs EMBL (European Molecular Biology Laboratory nucleotide sequence database at EBI, Hinxton, UK) Gen. Bank (at National Center for Biotechnology information, NCBI, Bethesda, MD, USA) DDBJ (DNA Data Bank Japan at CIB , Mishima, Japan) André de Carvalho - ICMC/USP 17/03/2018
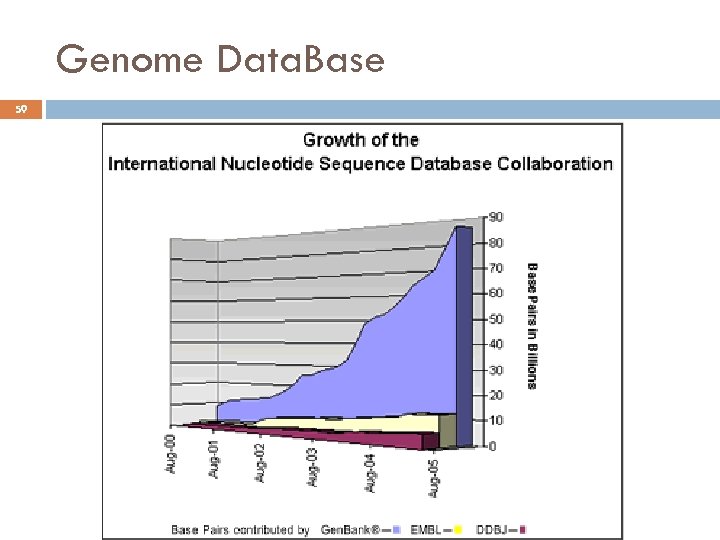
Genome Data. Base 59 André de Carvalho - ICMC/USP 17/03/2018
Gen. Bank 60 Each sequence Is identified by a unique adhesion number Includes a quantity of meta data Data about data or annotation Ex. : specie of the sequenced organism André de Carvalho - ICMC/USP 3/17/2018
Data Format and Annotation 61 There are several different formats to provide a sequence and its annotation EMBL, Gen. Bank and DDBJ have their own standard format There also formats that are not associated to a DB Generally to a sequence analyses program though FASTA André de Carvalho - ICMC/USP 3/17/2018
FASTA Format 62 André de Carvalho - ICMC/USP 17/03/2018
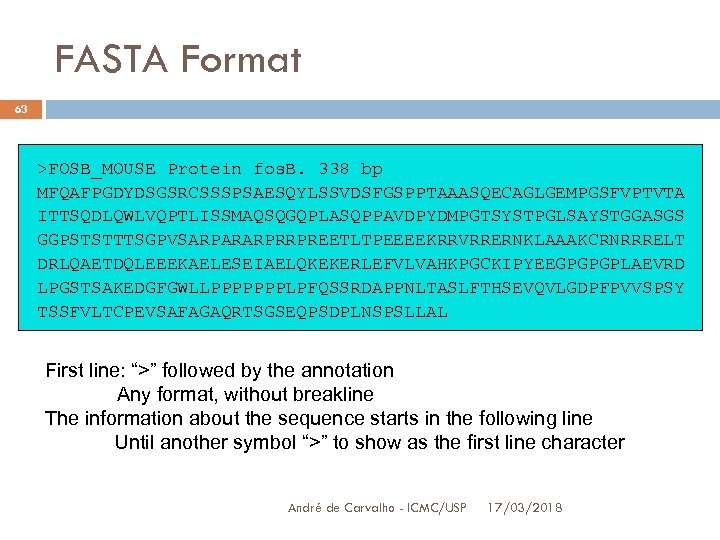
FASTA Format 63 >FOSB_MOUSE Protein fos. B. 338 bp MFQAFPGDYDSGSRCSSSPSAESQYLSSVDSFGSPPTAAASQECAGLGEMPGSFVPTVTA ITTSQDLQWLVQPTLISSMAQSQGQPLASQPPAVDPYDMPGTSYSTPGLSAYSTGGASGS GGPSTSTTTSGPVSARPARARPRRPREETLTPEEEEKRRVRRERNKLAAAKCRNRRRELT DRLQAETDQLEEEKAELESEIAELQKEKERLEFVLVAHKPGCKIPYEEGPGPGPLAEVRD LPGSTSAKEDGFGWLLPPPPPPPLPFQSSRDAPPNLTASLFTHSEVQVLGDPFPVVSPSY TSSFVLTCPEVSAFAGAQRTSGSEQPSDPLNSPSLLAL First line: “>” followed by the annotation Any format, without breakline The information about the sequence starts in the following line Until another symbol “>” to show as the first line character André de Carvalho - ICMC/USP 17/03/2018
FASTA Format 64 Accepted by the majority of the sequence analyses programs Provided by the majority of the online DBs Limits the amount of allowed annotation Other patterns are used to include more meta information Information about the sequence André de Carvalho - ICMC/USP 3/17/2018
Gen. Bank Format 65 An entry has several sections LOCUS: identifies the sequence DEFINITION: define the sequence ACCESSION: only identifies the sequence Related in publications and used to cross reference to other DBs SOURCE and ORGANISM: identifiy biological origin of the sequece REFERENCE: lists articles related to the sequence ORIGIN: lists all the nucleotides Among others André de Carvalho - ICMC/USP 17/03/2018
Gen. Bank Format 66 ORIGIN Sequences are organized in lines content 6 blocks, each of them with 10 bases Simbol “//” indicates the entry end André de Carvalho - ICMC/USP 17/03/2018
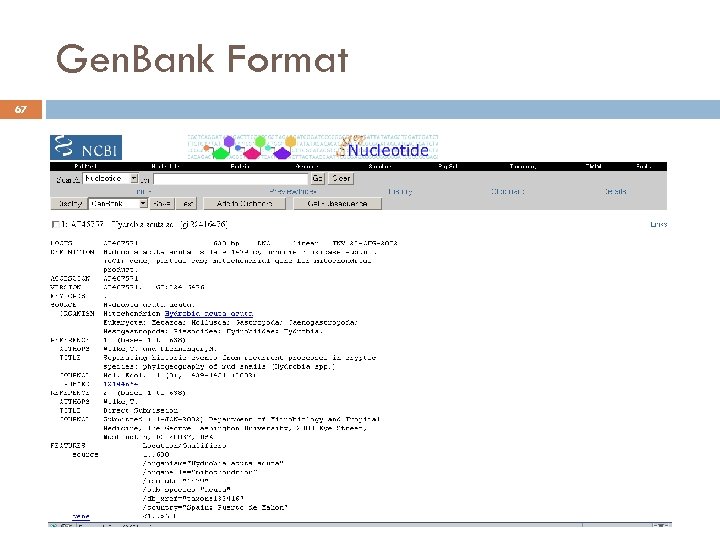
Gen. Bank Format 67 André de Carvalho - ICMC/USP 17/03/2018
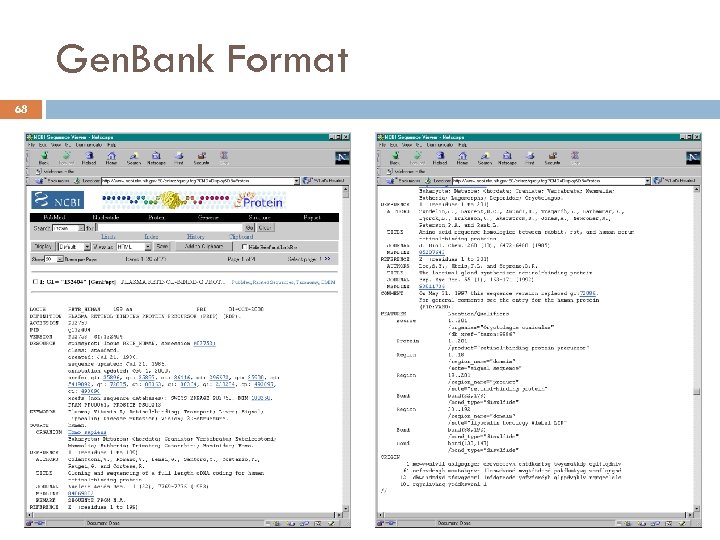
Gen. Bank Format 68 17/03/2018
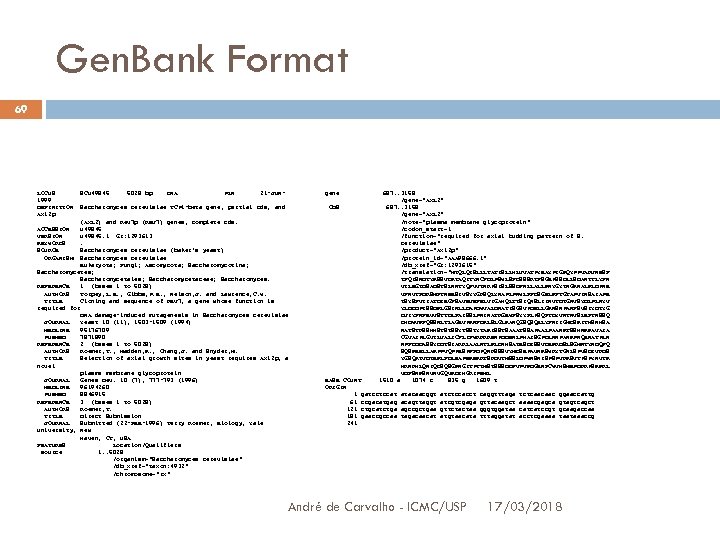
Gen. Bank Format 69 LOCUS 1999 DEFINITION Axl 2 p SCU 49845 5028 bp DNA PLN 21 -JUN- gene Saccharomyces cerevisiae TCP 1 -beta gene, partial cds, and CDS (AXL 2) and Rev 7 p (REV 7) genes, complete cds. ACCESSION U 49845 VERSION U 49845. 1 GI: 1293613 KEYWORDS. SOURCE Saccharomyces cerevisiae (baker's yeast) ORGANISM Saccharomyces cerevisiae Eukaryota; Fungi; Ascomycota; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Saccharomyces. REFERENCE 1 (bases 1 to 5028) AUTHORS Torpey, L. E. , Gibbs, P. E. , Nelson, J. and Lawrence, C. W. TITLE Cloning and sequence of REV 7, a gene whose function is required for DNA damage-induced mutagenesis in Saccharomyces cerevisiae JOURNAL Yeast 10 (11), 1503 -1509 (1994) MEDLINE 95176709 PUBMED 7871890 REFERENCE 2 (bases 1 to 5028) AUTHORS Roemer, T. , Madden, K. , Chang, J. and Snyder, M. TITLE Selection of axial growth sites in yeast requires Axl 2 p, a novel plasma membrane glycoprotein JOURNAL Genes Dev. 10 (7), 777 -793 (1996) MEDLINE 96194260 PUBMED 8846915 REFERENCE 3 (bases 1 to 5028) AUTHORS Roemer, T. TITLE Direct Submission JOURNAL Submitted (22 -FEB-1996) Terry Roemer, Biology, Yale University, New Haven, CT, USA FEATURES Location/Qualifiers source 1. . 5028 /organism="Saccharomyces cerevisiae" /db_xref="taxon: 4932" /chromosome="IX" 687. . 3158 /gene="AXL 2" /note="plasma membrane glycoprotein" /codon_start=1 /function="required for axial budding pattern of S. cerevisiae" /product="Axl 2 p" /protein_id="AAA 98666. 1" /db_xref="GI: 1293615" /translation="MTQLQISLLLTATISLLHLVVATPYEAYPIGKQYPPVARVNESF TFQISNDTYKSSVDKTAQITYNCFDLPSWLSFDSSSRTFSGEPSSDLLSDANTTLYFN VILEGTDSADSTSLNNTYQFVVTNRPSISLSSDFNLLALLKNYGYTNGKNALKLDPNE VFNVTFDRSMFTNEESIVSYYGRSQLYNAPLPNWLFFDSGELKFTGTAPVINSAIAPE TSYSFVIIATDIEGFSAVEVEFELVIGAHQLTTSIQNSLIINVTDTGNVSYDLPLNYV YLDDDPISSDKLGSINLLDAPDWVALDNATISGSVPDELLGKNSNPANFSVSIYDTYG DVIYFNFEVVSTTDLFAISSLPNINATRGEWFSYYFLPSQFTDYVNTNVSLEFTNSSQ DHDWVKFQSSNLTLAGEVPKNFDKLSLGLKANQGSQSQELYFNIIGMDSKITHSNHSA NATSTRSSHHSTSTSSYTSSTYTAKISSTSAAATSSAPAALPAANKTSSHNKKAVAIA CGVAIPLGVILVALICFLIFWRRRRENPDDENLPHAISGPDLNNPANKPNQENATPLN NPFDDDASSYDDTSIARRLAALNTLKLDNHSATESDISSVDEKRDSLSGMNTYNDQFQ SQSKEELLAKPPVQPPESPFFDPQNRSSSVYMDSEPAVNKSWRYTGNLSPVSDIVRDS YGSQKTVDTEKLFDLEAPEKEKRTSRDVTMSSLDPWNSNISPSPVRKSVTPSPYNVTK HRNRHLQNIQDSQSGKNGITPTTMSTSSSDDFVPVKDGENFCWVHSMEPDRRPSKKRL VDFSNKSNVNVGQVKDIHGRIPEML 1510 a 1074 c 835 g 1609 t BASE COUNT ORIGIN 1 gatcctccat 61 ccgacatgag 121 ctgcatctga 181 gaaccgccaa 241 atacaacggt acagttaggt agccgctgaa tagacaacat atctccacct atcgtcgaga gttctactaa atgtaacata André de Carvalho - ICMC/USP caggtttaga gttacaagct gggtggataa tttaggatat tctcaacaac aaaacgagca catcatccgt acctcgaaaa ggaaccattg gtagtcagct gcaagaccaa taataaaccg 17/03/2018
Pattern Alphabet 70 Sequences in different repositories follow the standard nucleotide alphabet Include symbols for ambiguous nucleotide Most common symbols: A Adenine C Citosine G Guanine T Thymine N any (a. Ny) base R A or G (pu. Rine) Y C or T (p. Yrimidine) M A or C (a. Mino) André de Carvalho - ICMC/USP 17/03/2018
Conclusion 71 Cells Molecular Biology Probabilistic Sequences Models Multinomial Models Markov Models Genome Annotation Data Base André de Carvalho - ICMC/USP 3/17/2018

Questions?

NCBI: National Center for Biotechnology information Established in 1988 as a national resource for molecular biology information, NCBI creates public databases, conducts research in computational biology, develops software tools for analyzing genome data, and disseminates biomedical information - all for the better understanding of molecular processes affecting human health and disease.

The EMBL Nucleotide Sequence Database (also known as EMBL-Bank) constitutes Europe's primary nucleotide sequence resource. Main sources for DNA and RNA sequences are direct submissions from individual researchers, genome sequencing projects and patent applications.
DDBJ (DNA Data Bank of Japan) began DNA data bank activities in earnest in 1986 at the National Institute of Genetics (NIG). DDBJ has been functioning as the international nucleotide sequence database in collaboration with EBI/EMBL and NCBI/Gen. Bank.
Fasta Protein Database Query Provides sequence similarity searching against nucleotide and protein databases using the Fasta programs. Fasta can be very specific when identifying long regions of low similarity especially for highly diverged sequences. You can also conduct sequence similarity searching against complete proteome or genome databases using the Fasta programs. Download Software
c9917b2efd17b48e8b7a6f389f254131.ppt