310d844d79c7d75b8423207556243a64.ppt
- Количество слайдов: 21

Sequence Alignment Gary Jackoway February 26, 2002 CISC 889: Bioinformatics February 26, 2002 Sequence Alignment -- Gary Jackoway 1
Sequence Alignment Outline n n n Dynamic Programming for Sequence Alignment Equivalent Problems Algorithm Description O(M*N) Proof By Example Global versus Local Alignment Nucleotide Substitution Matrix February 26, 2002 Sequence Alignment -- Gary Jackoway 2
Sequence Alignment Outline (cont) n n n PAM Substitution Matrix BLOSUM Substitution Matrix Log Odds Form Gap Penalty Alignment Issues Summary February 26, 2002 Sequence Alignment -- Gary Jackoway 3
Dynamic Programming for Sequence Alignment Problem: What is the “optimal” alignment of two DNA sequences. Input: Two DNA sequences (either Nucleotides or Amino Acids). Output: An “alignment” (mapping one sequence onto the other, possibly with gaps); and a “score” which defines the quality of the match. February 26, 2002 Sequence Alignment -- Gary Jackoway 4
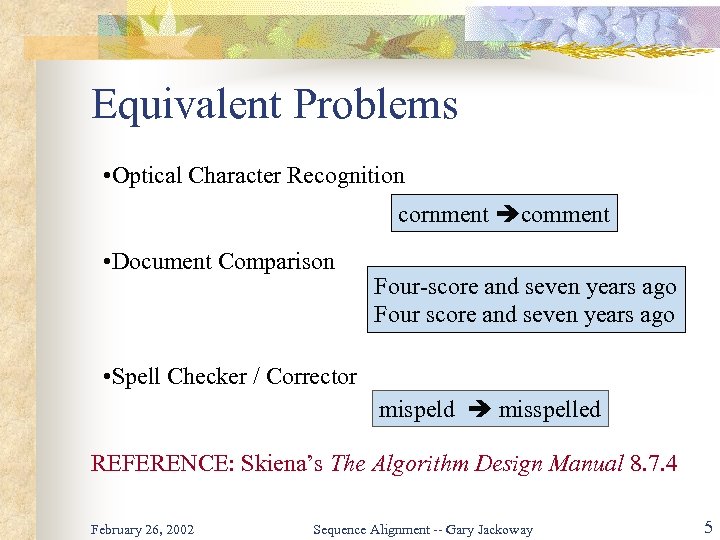
Equivalent Problems • Optical Character Recognition cornment comment • Document Comparison Four-score and seven years ago Four score and seven years ago • Spell Checker / Corrector mispeld misspelled REFERENCE: Skiena’s The Algorithm Design Manual 8. 7. 4 February 26, 2002 Sequence Alignment -- Gary Jackoway 5

Algorithm Description DP algorithms have a strong relationship to recursion: define a base case and prove that you can extend. If you already have the optimal solution to: X…Y A…B then you know the next pair of characters will either be: X…YZ or X…Y- or X…YZ A…BC A…B(where “-” indicates a gap). So you can extend the match by determining which of these has the highest score. February 26, 2002 Sequence Alignment -- Gary Jackoway 6
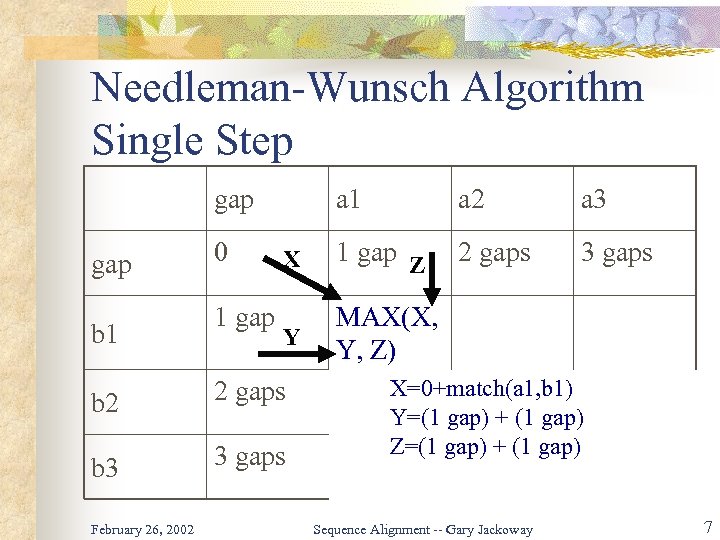
Needleman-Wunsch Algorithm Single Step gap b 1 0 1 gap a 1 a 2 a 3 X 1 gap Z 2 gaps 3 gaps Y MAX(X, Y, Z) b 2 2 gaps b 3 3 gaps February 26, 2002 X=0+match(a 1, b 1) Y=(1 gap) + (1 gap) Z=(1 gap) + (1 gap) Sequence Alignment -- Gary Jackoway 7
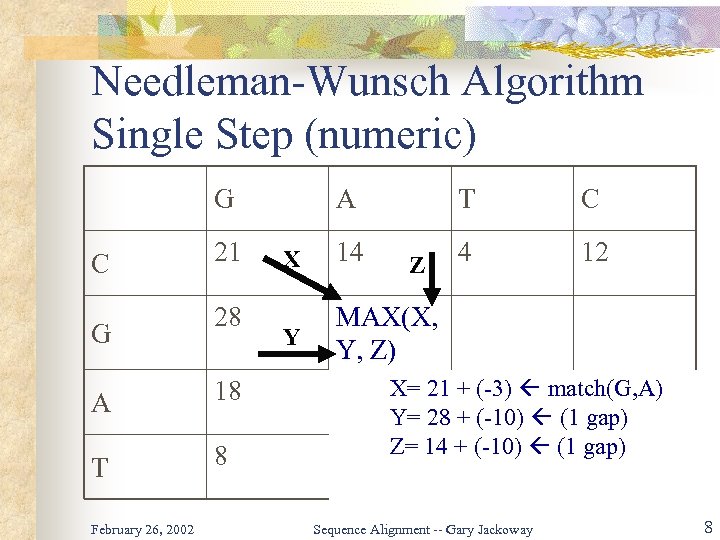
Needleman-Wunsch Algorithm Single Step (numeric) G C G 21 28 A 18 T 8 February 26, 2002 A T X 14 Y C 4 12 MAX(X, Y, Z) Z X= 21 + (-3) match(G, A) Y= 28 + (-10) (1 gap) Z= 14 + (-10) (1 gap) Sequence Alignment -- Gary Jackoway 8

O(M*N) Proof By Example We will prove that the dynamic programming algorithm for sequence alignment can be executed in O(M*N) time, where M=length of first sequence N=length of second sequence February 26, 2002 Sequence Alignment -- Gary Jackoway 9

Global versus Local Alignment Want to find local matching areas, even when far removed from each other in the sequence: ACTTAGCAGACTAACGTAAC CCATGACTAACGGGACCTAC Smith-Waterman: Use Needleman-Wunsch but add: IF value<0, replace with 0 (and set backtrack to none). When matrix is complete, backtrack from all local maxima, creating local matching alignments. February 26, 2002 Sequence Alignment -- Gary Jackoway 10
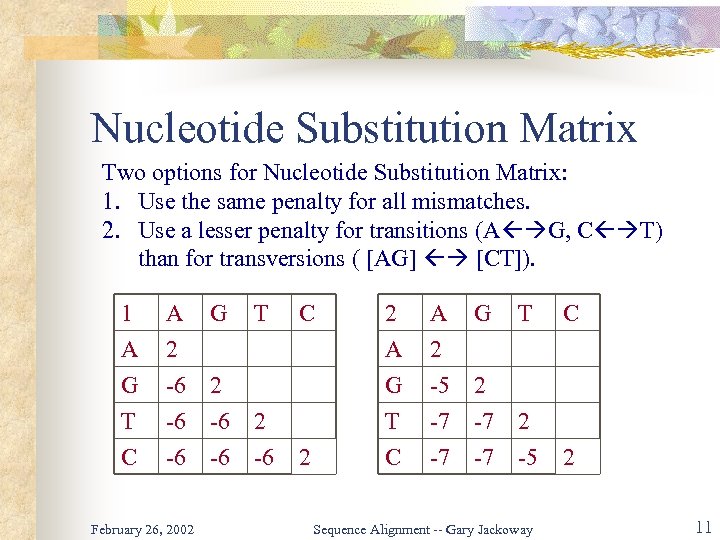
Nucleotide Substitution Matrix Two options for Nucleotide Substitution Matrix: 1. Use the same penalty for all mismatches. 2. Use a lesser penalty for transitions (A G, C T) than for transversions ( [AG] [CT]). 1 A G T A 2 -6 -6 G 2 -6 2 C -6 -6 -6 February 26, 2002 T C 2 2 A G T A 2 -5 -7 G T 2 -7 2 C -7 -7 -5 Sequence Alignment -- Gary Jackoway C 2 11
PAM: Percent Accepted Mutation Substitution Matrix (Dayhoff) n n n Substitution matrices based on sound evolutionary principles. Find PAM 1 by comparing groups of proteins known to be evolutionarily closely related. Find PAM-n my multiplying PAM 1 by itself n times. PAM 60: ~60% similar, PAM 250: ~20% similar. The more distant the expected relationship, the higher PAM-n should be used. February 26, 2002 Sequence Alignment -- Gary Jackoway 12
BLOSUM: BLOcks SUbstition Matrix n n Start with highly-conserved patterns (blocks) in a large set of closely related proteins. Use the likelihood of substitutions found in those sequences to create a substitution probability matrix. BLOSUM-n means that the sequences used were n% identical. BLOSUM 62 is “standard”. February 26, 2002 Sequence Alignment -- Gary Jackoway 13

Log Odds Form BLOSUM and PAM matrices start as a likelihood of substitution. Conversion to odds form yields a matrix that gives the odds that a change is evolutionarily significant versus purely random. Conversion to log odds form means that as you add each character to the pattern, you can add the values instead of multiplying them (as you would need to do for odds form). February 26, 2002 Sequence Alignment -- Gary Jackoway 14

Gap Penalty n The gap penalty has to “work” with the substitution matrix. (Ex. if you have a gap penalty that is not more severe than two substitutions, then you will get an insert / delete pair instead of a substitution. ) n n If gap penalty is too costly, will get mismatches when a gap would lead to a better match. If gap penalty is too cheap, will get meaningless gaps, just to line up one or two characters. February 26, 2002 Sequence Alignment -- Gary Jackoway 15

Gap Penalty (cont. ) n n It is intuitively appealing to use a gap penalty of the form g+r*x where x is the length of the gap, “r” is the “gap extension penalty”. It is better to have one big gap than scattered small ones. NOTE: If the gap penalty (or extension) is not more costly than all substitutions, the recurrence relation needs correction: need to look back along the current row and column to assure optimality. [Violates the “triangle inequality”. ] February 26, 2002 Sequence Alignment -- Gary Jackoway 16

How good is my alignment? (Starting with log odds form helps. ) Most online programs give a number of statistical formulations that attempt to answer the question. score: the value calculated for the sequence using the substitution matrix and the gap penalties. percent identity: percent of exact matching symbols. Expected value (E): probability that a match with this score would be obtained comparing two random sequences. NOTE: different systems use different forms of this statistic. February 26, 2002 Sequence Alignment -- Gary Jackoway 17

Alignment Questions Should I use a global or a local alignment algorithm? Which substitution matrix should I use? What gap penalty structure should I use? The answer to all of these questions lies in your response to this question: What are you trying to find out? February 26, 2002 Sequence Alignment -- Gary Jackoway 18

What are you trying to find out? n n n Are you trying to locate similar domains or motifs? Local alignment is probably best. Are you trying to determine whether the sequences are from the same family? Use one of the BLOSUM matrices. Are you trying to determine how closely related the sequences are evolutionarily? Use one of the PAM matrices. February 26, 2002 Sequence Alignment -- Gary Jackoway 19
Summary n n n Sequence Alignment is a powerful tool for determining relatedness between two sequences. There are many options and decisions to make in determining how to do the alignment. It is essential to understand what type of relationship one is looking for in order to apply the right tool with the right parameter set. February 26, 2002 Sequence Alignment -- Gary Jackoway 20

Summary (cont) n Online resources can be found in table 3. 1 of the book or www. bioinformaticsonline. org. Recommend: BCM-SIM, BCM-BLAST 2, FASTALALIGN, FASTA-PRSS, BLAST 2 n n Another interesting resource is the Genome Multimedia Site: ocelot. bio. brandeis. edu / pages/classes/Interp. Genes/Project/menu. htm Never underestimate the power of a good spreadsheet! February 26, 2002 Sequence Alignment -- Gary Jackoway 21
310d844d79c7d75b8423207556243a64.ppt