7f88a376fc9a6eae53cbb2b0cbe5738b.ppt
- Количество слайдов: 16

Semantic Web Technologies for Translational Medicine Vipul Kashyap, Ph. D vkashyap 1@partners. org Senior Medical Informatician, Clinical Knowledge Management and Decision Support Clinical Informatics R&D, Partners Healthcare System Panel on “Towards a Semantic Web for the Life Sciences? ” October 24, 2005

Outline • Translational Medicine Use Case — Translation of Genomic Research Insights into Clinical Care • Key Functionalities — Data Integration — Actionable Decision Support — Knowledge Update and Propagation • Semantic Web Technologies — RDF: Resource Description Framework — OWL: Web Ontology Language — SWRL: Semantic Web Rules Language • Conclusions
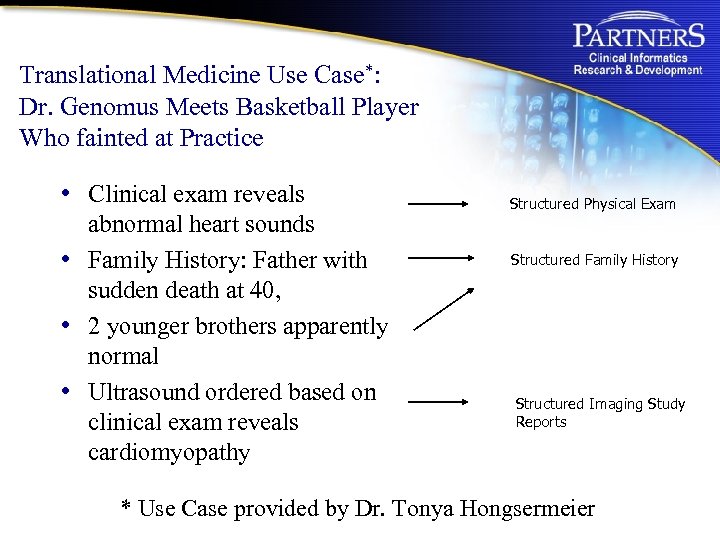
Translational Medicine Use Case*: Dr. Genomus Meets Basketball Player Who fainted at Practice • Clinical exam reveals abnormal heart sounds • Family History: Father with sudden death at 40, • 2 younger brothers apparently normal • Ultrasound ordered based on clinical exam reveals cardiomyopathy Structured Physical Exam Structured Family History Structured Imaging Study Reports * Use Case provided by Dr. Tonya Hongsermeier
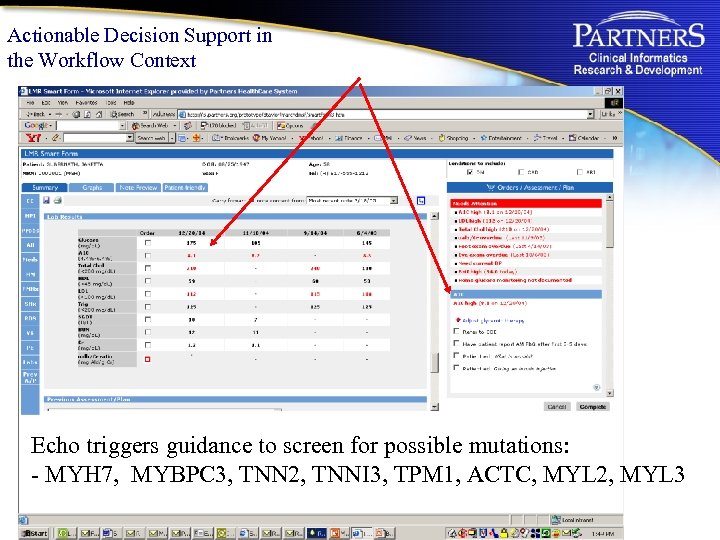
Actionable Decision Support in the Workflow Context Echo triggers guidance to screen for possible mutations: - MYH 7, MYBPC 3, TNN 2, TNNI 3, TPM 1, ACTC, MYL 2, MYL 3
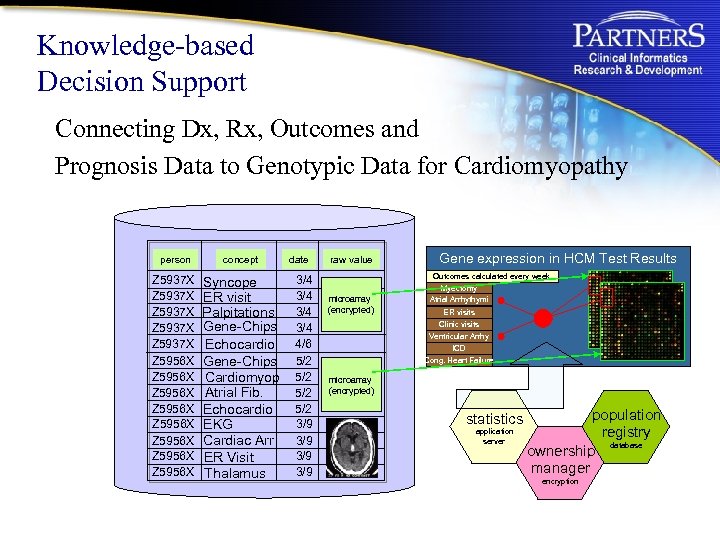
Knowledge-based Decision Support Connecting Dx, Rx, Outcomes and Prognosis Data to Genotypic Data for Cardiomyopathy person concept Z 5937 X Z 5937 X Z 5956 X Z 5956 X Syncope ER visit Palpitations Gene-Chips Echocardio Gene-Chips Cardiomyop Atrial Fib. Echocardio EKG Cardiac Arr ER Visit Thalamus date 3/4 3/4 4/6 5/2 5/2 3/9 3/9 raw value microarray (encrypted) Gene expression in HCM Test Results Outcomes calculated every week Myectomy Atrial Arrhythymi ER visits Clinic visits Ventricular Arrhy ICD Cong. Heart Failure microarray (encrypted) statistics application server population registry database ownership manager encryption
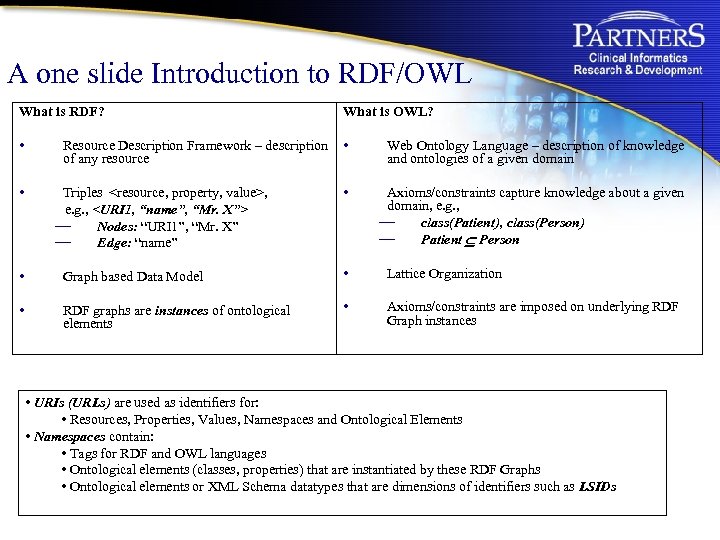
A one slide Introduction to RDF/OWL What is RDF? What is OWL? • • Web Ontology Language – description of knowledge and ontologies of a given domain • Axioms/constraints capture knowledge about a given domain, e. g. , — class(Patient), class(Person) — Patient Person • Resource Description Framework – description of any resource Triples <resource, property, value>, e. g. , <URI 1, “name”, “Mr. X”> — Nodes: “URI 1”, “Mr. X” — Edge: “name” • Graph based Data Model • Lattice Organization • RDF graphs are instances of ontological elements • Axioms/constraints are imposed on underlying RDF Graph instances • URIs (URLs) are used as identifiers for: • Resources, Properties, Values, Namespaces and Ontological Elements • Namespaces contain: • Tags for RDF and OWL languages • Ontological elements (classes, properties) that are instantiated by these RDF Graphs • Ontological elements or XML Schema datatypes that are dimensions of identifiers such as LSIDs
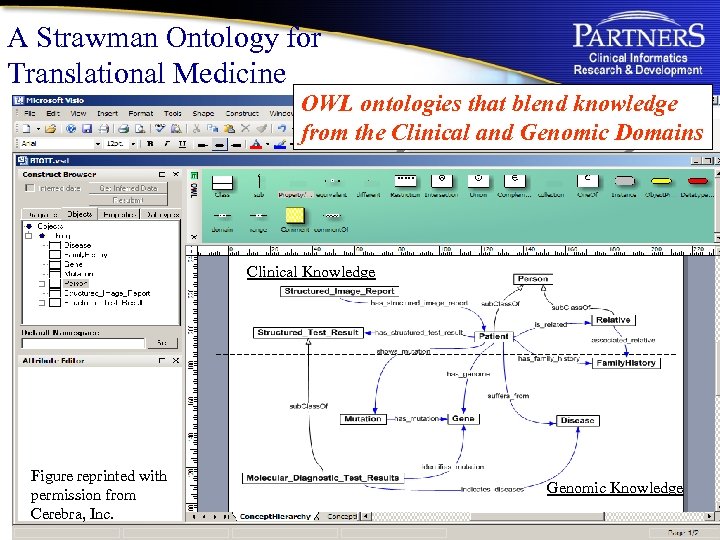
A Strawman Ontology for Translational Medicine OWL ontologies that blend knowledge from the Clinical and Genomic Domains Clinical Knowledge Figure reprinted with permission from Cerebra, Inc. Genomic Knowledge
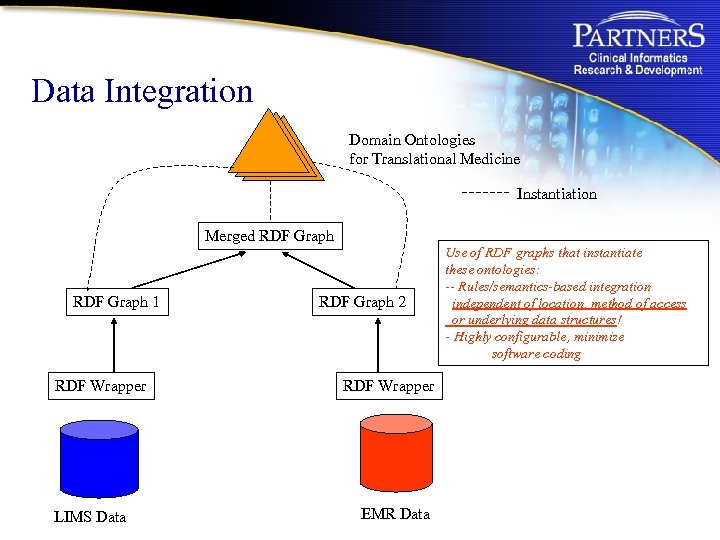
Data Integration Domain Ontologies for Translational Medicine Instantiation Merged RDF Graph 1 RDF Wrapper LIMS Data RDF Graph 2 RDF Wrapper EMR Data Use of RDF graphs that instantiate these ontologies: -- Rules/semantics-based integration independent of location, method of access or underlying data structures! - Highly configurable, minimize software coding
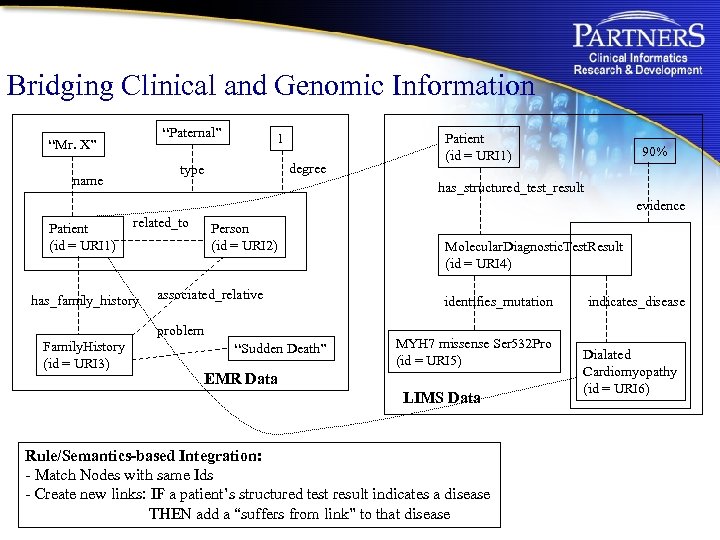
Bridging Clinical and Genomic Information “Paternal” “Mr. X” 1 degree type name Patient (id = URI 1) 90% has_structured_test_result evidence Patient (id = URI 1) related_to has_family_history Person (id = URI 2) associated_relative problem Family. History (id = URI 3) “Sudden Death” Molecular. Diagnostic. Test. Result (id = URI 4) identifies_mutation MYH 7 missense Ser 532 Pro (id = URI 5) EMR Data LIMS Data Rule/Semantics-based Integration: - Match Nodes with same Ids - Create new links: IF a patient’s structured test result indicates a disease THEN add a “suffers from link” to that disease indicates_disease Dialated Cardiomyopathy (id = URI 6)
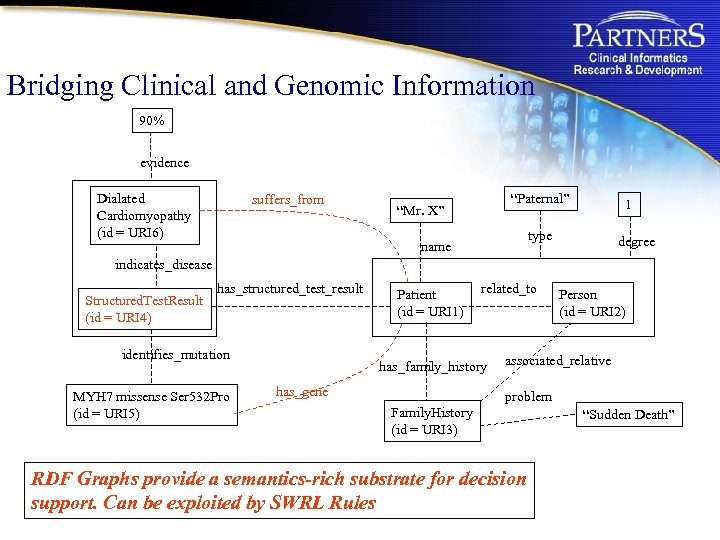
Bridging Clinical and Genomic Information 90% evidence Dialated Cardiomyopathy (id = URI 6) suffers_from “Paternal” “Mr. X” 1 type name degree indicates_disease Structured. Test. Result (id = URI 4) has_structured_test_result identifies_mutation MYH 7 missense Ser 532 Pro (id = URI 5) Patient (id = URI 1) related_to has_family_history has_gene Person (id = URI 2) associated_relative problem Family. History (id = URI 3) RDF Graphs provide a semantics-rich substrate for decision support. Can be exploited by SWRL Rules “Sudden Death”
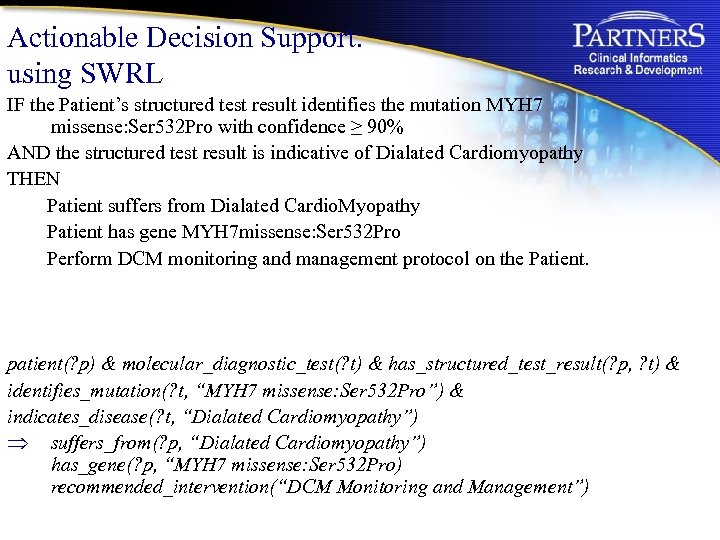
Actionable Decision Support: using SWRL IF the Patient’s structured test result identifies the mutation MYH 7 missense: Ser 532 Pro with confidence ≥ 90% AND the structured test result is indicative of Dialated Cardiomyopathy THEN Patient suffers from Dialated Cardio. Myopathy Patient has gene MYH 7 missense: Ser 532 Pro Perform DCM monitoring and management protocol on the Patient. patient(? p) & molecular_diagnostic_test(? t) & has_structured_test_result(? p, ? t) & identifies_mutation(? t, “MYH 7 missense: Ser 532 Pro”) & indicates_disease(? t, “Dialated Cardiomyopathy”) Þ suffers_from(? p, “Dialated Cardiomyopathy”) has_gene(? p, “MYH 7 missense: Ser 532 Pro) recommended_intervention(“DCM Monitoring and Management”)

Semantic Web Rules Language (SWRL) • • • References to ontological concepts and relationships — Describe clinical and genomic information Can be used to infer patient state: — Patient has a particular gene/mutation — Patient suffers from a particular disease Can be used to recommend clinical care: — Order Monitoring and Management Protocol patient(? p) & molecular_diagnostic_test(? t) & mutation(? m) & disease(? d) has_structured_test_result(? p, ? t) & identifies_mutation(? t, ? m) & indicates_disease(? t, ? d) & suggested_protocol(? d, ? pro) Þ suffers_from(? p, ? d) has_gene(? p, ? m) order_protocol(? pro)
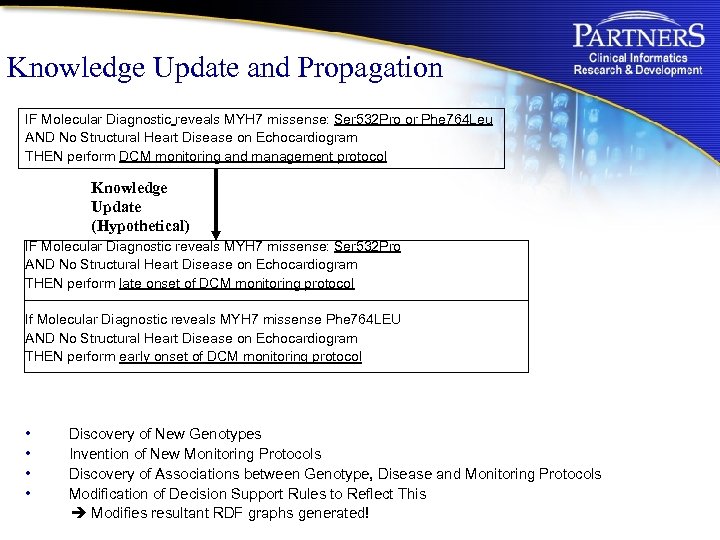
Knowledge Update and Propagation IF Molecular Diagnostic reveals MYH 7 missense: Ser 532 Pro or Phe 764 Leu AND No Structural Heart Disease on Echocardiogram THEN perform DCM monitoring and management protocol Knowledge Update (Hypothetical) IF Molecular Diagnostic reveals MYH 7 missense: Ser 532 Pro AND No Structural Heart Disease on Echocardiogram THEN perform late onset of DCM monitoring protocol If Molecular Diagnostic reveals MYH 7 missense Phe 764 LEU AND No Structural Heart Disease on Echocardiogram THEN perform early onset of DCM monitoring protocol • • Discovery of New Genotypes Invention of New Monitoring Protocols Discovery of Associations between Genotype, Disease and Monitoring Protocols Modification of Decision Support Rules to Reflect This Modifies resultant RDF graphs generated!
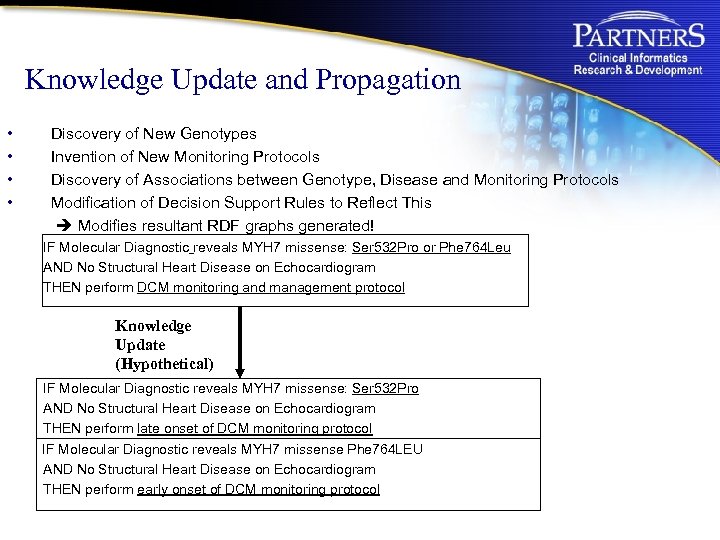
Knowledge Update and Propagation • • Discovery of New Genotypes Invention of New Monitoring Protocols Discovery of Associations between Genotype, Disease and Monitoring Protocols Modification of Decision Support Rules to Reflect This Modifies resultant RDF graphs generated! IF Molecular Diagnostic reveals MYH 7 missense: Ser 532 Pro or Phe 764 Leu AND No Structural Heart Disease on Echocardiogram THEN perform DCM monitoring and management protocol Knowledge Update (Hypothetical) IF Molecular Diagnostic reveals MYH 7 missense: Ser 532 Pro AND No Structural Heart Disease on Echocardiogram THEN perform late onset of DCM monitoring protocol IF Molecular Diagnostic reveals MYH 7 missense Phe 764 LEU AND No Structural Heart Disease on Echocardiogram THEN perform early onset of DCM monitoring protocol
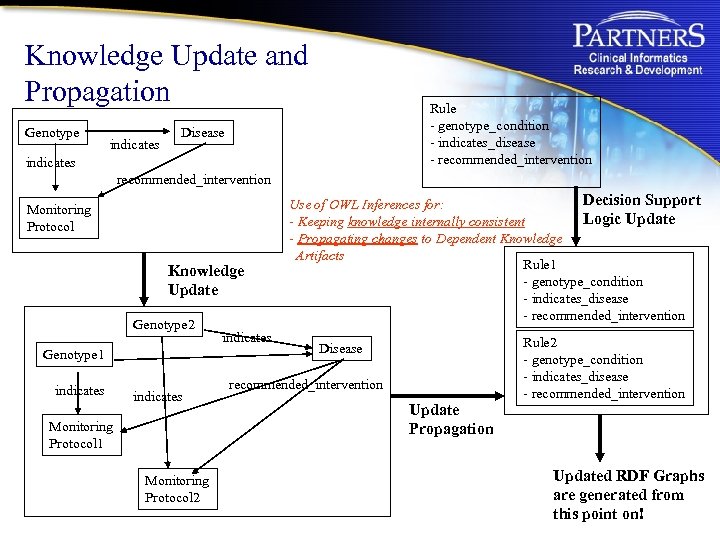
Knowledge Update and Propagation Genotype indicates Rule - genotype_condition - indicates_disease - recommended_intervention Disease indicates recommended_intervention Monitoring Protocol Knowledge Update Genotype 2 Genotype 1 indicates Monitoring Protocol 1 Monitoring Protocol 2 indicates Decision Support Use of OWL Inferences for: Logic Update - Keeping knowledge internally consistent - Propagating changes to Dependent Knowledge Artifacts Rule 1 - genotype_condition - indicates_disease - recommended_intervention Rule 2 - genotype_condition - indicates_disease - recommended_intervention Disease recommended_intervention Update Propagation Updated RDF Graphs are generated from this point on!

Conclusions • Translational Medicine is a knowledge intensive field. The ability to capture semantics of this knowledge is crucial for implementation. • Personalized Medicine cannot be implemented in an scalable, efficient and extensible manner without Semantic Web technologies • The rate of Knowledge Updates will change drastically as Genomic knowledge explodes • Automated Semantics-based Knowledge Update and Propagation will be key in keeping the knowledge updated and current
7f88a376fc9a6eae53cbb2b0cbe5738b.ppt