2d092675a067ebea028a1afa0b25b1e0.ppt
- Количество слайдов: 14
 Running ORGANON in R Peter Gould and David Marshall Growth Model User Group December 9, 2011
Running ORGANON in R Peter Gould and David Marshall Growth Model User Group December 9, 2011
 Why run ORGANON in R? • R is an excellent environment for analysis. • You can do just about any computing task in R. • Connects with different data formats (text files, Access databases, SQL databases). • Produced excellent graphics, also tables and other types of text output. • Many tasks can be automated. • If you can do it in R, why do it any other way?
Why run ORGANON in R? • R is an excellent environment for analysis. • You can do just about any computing task in R. • Connects with different data formats (text files, Access databases, SQL databases). • Produced excellent graphics, also tables and other types of text output. • Many tasks can be automated. • If you can do it in R, why do it any other way?
 What do we need to get started? • Computer with Windows OS • R (free download from website). • ORGANON source code (download from website). • Freeware FORTRAN compiler (gfortran, installed as part of Min. GW). • A little patience…
What do we need to get started? • Computer with Windows OS • R (free download from website). • ORGANON source code (download from website). • Freeware FORTRAN compiler (gfortran, installed as part of Min. GW). • A little patience…
 Everything’s installed, what do we do now? • Make a few changes to the ORGANON source code. • Recompile the dlls* using gfortran. • Write R scripts to access the dlls. *Dynamic link library: a library of subroutines that can be called by a Windows application.
Everything’s installed, what do we do now? • Make a few changes to the ORGANON source code. • Recompile the dlls* using gfortran. • Write R scripts to access the dlls. *Dynamic link library: a library of subroutines that can be called by a Windows application.
 Changes to the Source Code • Compiling with gfortran will make “Rcompatible” dlls but a few things need to be changed: • Remove or comment out all lines that contain “DLL EXPORT” in these files: • PREPARE. FOR • EXECUTE 2. FOR • ORGVOL. FOR • WOODQUAL. FOR • Change the functions JIFIX and INT 4 to LONG in: • ORGVOL. FOR • VOLEQNS. FOR
Changes to the Source Code • Compiling with gfortran will make “Rcompatible” dlls but a few things need to be changed: • Remove or comment out all lines that contain “DLL EXPORT” in these files: • PREPARE. FOR • EXECUTE 2. FOR • ORGVOL. FOR • WOODQUAL. FOR • Change the functions JIFIX and INT 4 to LONG in: • ORGVOL. FOR • VOLEQNS. FOR
 You can also make other changes. Example: add a subroutine to identify the new dll: C RECORD CHANGES WHEN THE DLL IS RECOMPILED SUBROUTINE REVISION(HISTORY) CHARACTER*500 HISTORY='Recompiled by Peter Gould Nov 13, 2011. Minor changes' 1 //'were made for compatibility with gfortran. No substantive' 2 //' changes were made. Contact: pgould@fs. fed. us' END
You can also make other changes. Example: add a subroutine to identify the new dll: C RECORD CHANGES WHEN THE DLL IS RECOMPILED SUBROUTINE REVISION(HISTORY) CHARACTER*500 HISTORY='Recompiled by Peter Gould Nov 13, 2011. Minor changes' 1 //'were made for compatibility with gfortran. No substantive' 2 //' changes were made. Contact: pgould@fs. fed. us' END
 Compile the dlls using gfortran • Compiler is called from the command prompt. gfortran -shared -static-libgcc -o ORGEDIT. dll DIAMCAL. FOR COMFILES. FOR START 2. FOR PREPARE. FOR • Edit the PATH variable to make the call to the compiler easier.
Compile the dlls using gfortran • Compiler is called from the command prompt. gfortran -shared -static-libgcc -o ORGEDIT. dll DIAMCAL. FOR COMFILES. FOR START 2. FOR PREPARE. FOR • Edit the PATH variable to make the call to the compiler easier.
 We can also do all these things with R … Download Unzip files Make needed changes Compile ###Download and compile ORGANON dlls ##Peter Gould Dec 6, 2011 ###set a working directory setwd("C: /Adata. Folder/ORGANON/R_EXAMPLE") ###download ORGANON download. file("http: //www. cof. orst. edu/cof/fr/research/organon/ORGANON%20 DLLS%20 SOURCE%20 CODE. ZIP", "ORGSOURCE. ZIP", mode="wb") ##unzip file unzip("ORGSOURCE. ZIP") unzip("EDITDLL SOURCE CODE. ZIP") unzip("RUNDLL SOURCE CODE. ZIP") unzip("VOLDLL SOURCE CODE. ZIP") unzip("WQDLL SOURCE CODE. ZIP") ###edit files allfiles = dir(pattern=". FOR", recursive = T, full. name=T) for(getfile in allfiles){ read 1 = read. table(getfile, sep="~", as. is=T, quote="") read 1 = read 1$V 1 ##remove DLL statements read 1 = read 1[!grepl("DLL_EXPORT", read 1)] ##replace JIFIX statements read 1 =sub("JIFIX", "LONG", read 1) ##replace INT 4 statements read 1 =sub("INT 4", "LONG", read 1) ##re-write the file write(read 1, getfile, ncolumns=1) } ##write a batch file to run the compiler send = rep("", 5) send[1] = getwd() send[1] = gsub("/", "\\", send[1]) send[1] = paste("cd", send[1]) send[2] = "gfortran -shared -static-libgcc -o ORGEDIT. dll DIAMCAL. FOR COMFILES. FOR START 2. FOR PREPARE. FOR" send[3] = "gfortran -shared -static-libgcc -static-libgfortran -o ORGRUN. dll CRNGROW. FOR DIAGRO. FOR EXECUTE 2. FOR GROWTH_MODS. FOR HTGROWTH. FOR MORTALITY. FOR STATS. FOR SUBMAX. FOR TRIPLE. FOR WHPHG. FOR" send[4] = "gfortran -shared -static-libgcc -o ORGVOL. dll ORGVOL. FOR VOLEQNS. FOR" send[5] = "gfortran -shared -static-libgcc -o ORGWQ. dll COMFILES 2. FOR WOODQUAL. FOR" write. table(send, "COMPILE. BAT", row. names=F, col. names=F, quote=F) system("COMPILE. BAT")
We can also do all these things with R … Download Unzip files Make needed changes Compile ###Download and compile ORGANON dlls ##Peter Gould Dec 6, 2011 ###set a working directory setwd("C: /Adata. Folder/ORGANON/R_EXAMPLE") ###download ORGANON download. file("http: //www. cof. orst. edu/cof/fr/research/organon/ORGANON%20 DLLS%20 SOURCE%20 CODE. ZIP", "ORGSOURCE. ZIP", mode="wb") ##unzip file unzip("ORGSOURCE. ZIP") unzip("EDITDLL SOURCE CODE. ZIP") unzip("RUNDLL SOURCE CODE. ZIP") unzip("VOLDLL SOURCE CODE. ZIP") unzip("WQDLL SOURCE CODE. ZIP") ###edit files allfiles = dir(pattern=". FOR", recursive = T, full. name=T) for(getfile in allfiles){ read 1 = read. table(getfile, sep="~", as. is=T, quote="") read 1 = read 1$V 1 ##remove DLL statements read 1 = read 1[!grepl("DLL_EXPORT", read 1)] ##replace JIFIX statements read 1 =sub("JIFIX", "LONG", read 1) ##replace INT 4 statements read 1 =sub("INT 4", "LONG", read 1) ##re-write the file write(read 1, getfile, ncolumns=1) } ##write a batch file to run the compiler send = rep("", 5) send[1] = getwd() send[1] = gsub("/", "\\", send[1]) send[1] = paste("cd", send[1]) send[2] = "gfortran -shared -static-libgcc -o ORGEDIT. dll DIAMCAL. FOR COMFILES. FOR START 2. FOR PREPARE. FOR" send[3] = "gfortran -shared -static-libgcc -static-libgfortran -o ORGRUN. dll CRNGROW. FOR DIAGRO. FOR EXECUTE 2. FOR GROWTH_MODS. FOR HTGROWTH. FOR MORTALITY. FOR STATS. FOR SUBMAX. FOR TRIPLE. FOR WHPHG. FOR" send[4] = "gfortran -shared -static-libgcc -o ORGVOL. dll ORGVOL. FOR VOLEQNS. FOR" send[5] = "gfortran -shared -static-libgcc -o ORGWQ. dll COMFILES 2. FOR WOODQUAL. FOR" write. table(send, "COMPILE. BAT", row. names=F, col. names=F, quote=F) system("COMPILE. BAT")
 1. 2. 3. 4. Making a model run Load data into R. Format data to get it ready to run. Load the dlls. Call the subroutine “prepare” to fill-in heights and crown ratios. 5. Project the stand 1 cycle (5 yrs) by calling “execute”. 6. Load the projected values into the initial values. 7. Repeat steps 4 and 5 until the projection is completed.
1. 2. 3. 4. Making a model run Load data into R. Format data to get it ready to run. Load the dlls. Call the subroutine “prepare” to fill-in heights and crown ratios. 5. Project the stand 1 cycle (5 yrs) by calling “execute”. 6. Load the projected values into the initial values. 7. Repeat steps 4 and 5 until the projection is completed.
 Loading the dlls • dyn. load() ###load dll ##DEFINE THE DIRECTORY WHERE THE DLLS WERE PLACED setwd("C: /Adata. Folder/ORGANON/COMPILE") dyn. load("ORGEDIT. dll", type="Fortran") dyn. load("ORGRUN. dll", type="Fortran") dyn. load("ORGVOL. dll", type="Fortran")
Loading the dlls • dyn. load() ###load dll ##DEFINE THE DIRECTORY WHERE THE DLLS WERE PLACED setwd("C: /Adata. Folder/ORGANON/COMPILE") dyn. load("ORGEDIT. dll", type="Fortran") dyn. load("ORGRUN. dll", type="Fortran") dyn. load("ORGVOL. dll", type="Fortran")
 Calling a dll from R: • All variables must be initialized first: ## initialize variables VERSION = 1 NPTS = 2 NTREES = 10 STAGE = 40 BHAGE = 37 • Variables must be “cast” within a call: grow =. Fortran("execute", as. integer(CYCLEG), as. integer(VERSION), as. integer(NPTS), as. integer(NTREES 1), as. integer(STAGE), as. integer(BHAGE), as. integer(TREENO), as. integer(PTNO), as. integer(SPECIES), as. integer(USER), as. integer(INDS), as. single(DBH 1), as. single(HT 1), as. single(CR 1), as. single(SCR 1), as. single(EXPAN 1), as. single(MGEXP),
Calling a dll from R: • All variables must be initialized first: ## initialize variables VERSION = 1 NPTS = 2 NTREES = 10 STAGE = 40 BHAGE = 37 • Variables must be “cast” within a call: grow =. Fortran("execute", as. integer(CYCLEG), as. integer(VERSION), as. integer(NPTS), as. integer(NTREES 1), as. integer(STAGE), as. integer(BHAGE), as. integer(TREENO), as. integer(PTNO), as. integer(SPECIES), as. integer(USER), as. integer(INDS), as. single(DBH 1), as. single(HT 1), as. single(CR 1), as. single(SCR 1), as. single(EXPAN 1), as. single(MGEXP),
 Example Script Double-click icon to open. Doesn’t work in “slide show” mode.
Example Script Double-click icon to open. Doesn’t work in “slide show” mode.
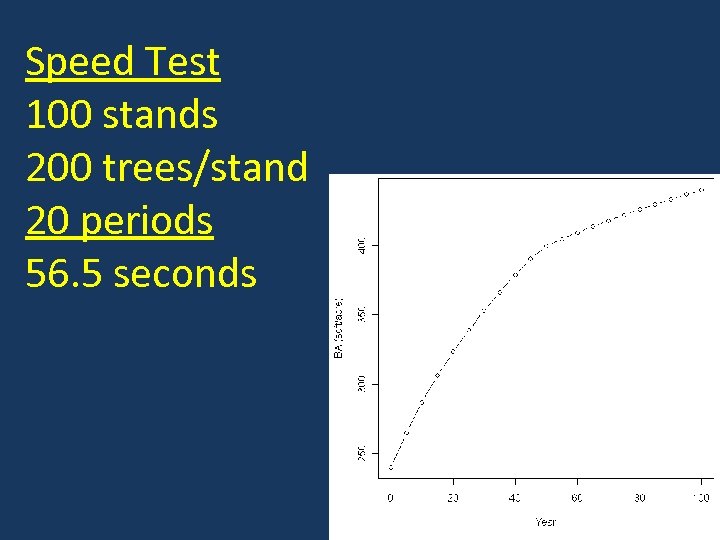 Speed Test 100 stands 200 trees/stand 20 periods 56. 5 seconds
Speed Test 100 stands 200 trees/stand 20 periods 56. 5 seconds
 What can we do now? • Create an R package for ORGANON. • Can also make individual functions available such as “volcal” and “prepare” to impute heights and crown ratios. • Create functions to “seamlessly” hand off projections from CONIFERS to ORGANON. • Create a working custom version of the main ORGANON growth model. • Do simple or elaborate projections/analyses within R.
What can we do now? • Create an R package for ORGANON. • Can also make individual functions available such as “volcal” and “prepare” to impute heights and crown ratios. • Create functions to “seamlessly” hand off projections from CONIFERS to ORGANON. • Create a working custom version of the main ORGANON growth model. • Do simple or elaborate projections/analyses within R.


