39904eb5a8510c001df6abfa091bbf23.ppt
- Количество слайдов: 30

Rule Extraction From Trained Neural Networks Brian Hudson University of Portsmouth, UK
Artificial Neural Networks Advantages High accuracy Robust Noisy data Disadvantages Lack of comprehensibilty
Trepan A method for extracting a decision tree from an artificial neural network (Craven, 1996). The tree is built by expanding nodes in a best first manner, producing an unbalanced tree. The splitting tests at the nodes are m-of-n tests e. g. 2 -of-{x 1, ¬x 2, x 3}, where the xi are Boolean conditions The network is used as an oracle to answer queries during the learning process.

Splitting Tests Start with a set of candidate tests binary tests on each value for nominal features binary tests on thresholds for real-valued features Find optimal splitting test by a beam search, initializing beam with candidate test maximizing the information gain.
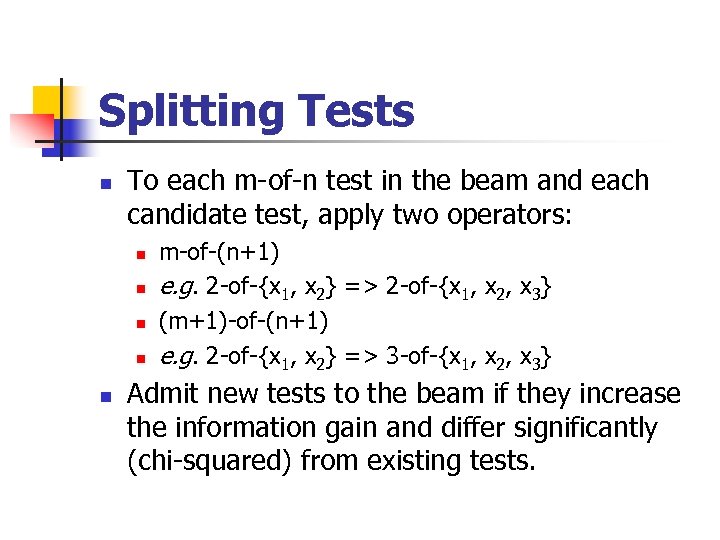
Splitting Tests To each m-of-n test in the beam and each candidate test, apply two operators: m-of-(n+1) e. g. 2 -of-{x 1, x 2} => 2 -of-{x 1, x 2, x 3} (m+1)-of-(n+1) e. g. 2 -of-{x 1, x 2} => 3 -of-{x 1, x 2, x 3} Admit new tests to the beam if they increase the information gain and differ significantly (chi-squared) from existing tests.
Data Modelling The amount of training data reaching each node decreases with depth of tree. TREPAN creates new training cases by sampling the distributions of the training data empirical distributions for nominal inputs kernel density estimates for continuous inputs Apply oracle (i. e. neural network) to new training cases to assign output values.

Application to Bioinformatics Prediction of Splice Junction sites in Eukaryotic DNA
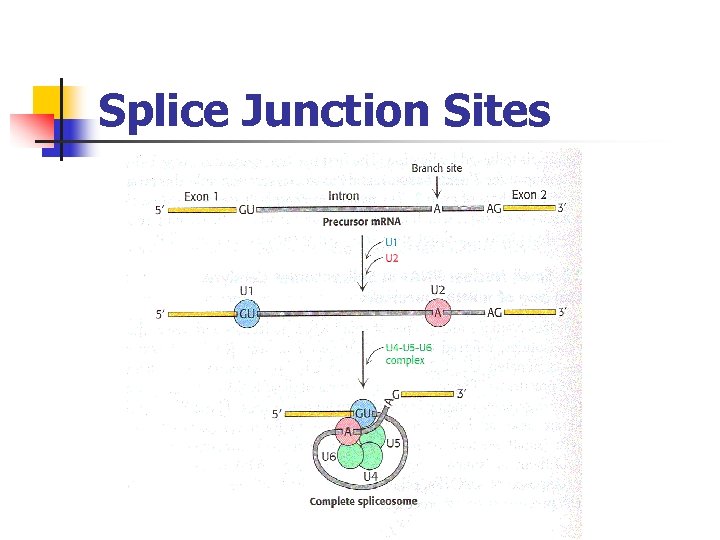
Splice Junction Sites
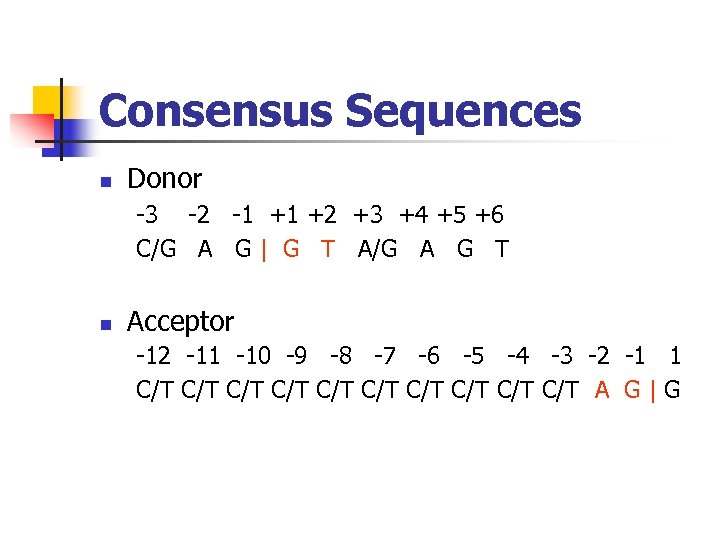
Consensus Sequences Donor -3 -2 -1 +1 +2 +3 +4 +5 +6 C/G A G | G T A/G A G T Acceptor -12 -11 -10 -9 -8 -7 -6 -5 -4 -3 -2 -1 1 C/T C/T C/T A G | G
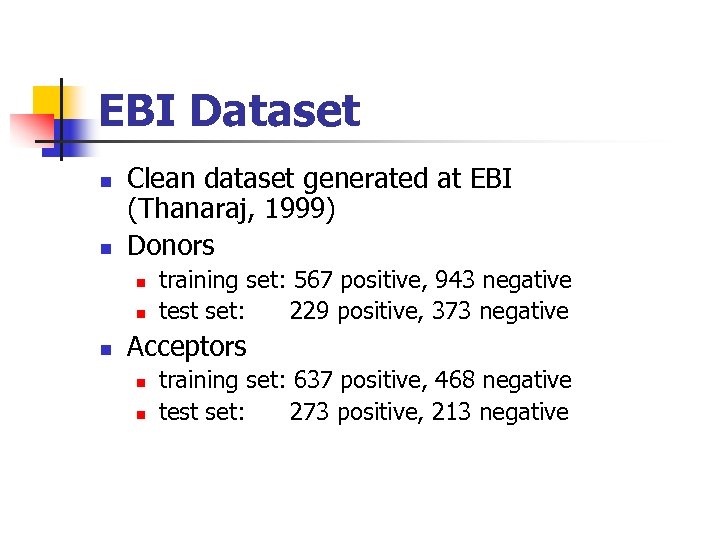
EBI Dataset Clean dataset generated at EBI (Thanaraj, 1999) Donors training set: 567 positive, 943 negative test set: 229 positive, 373 negative Acceptors training set: 637 positive, 468 negative test set: 273 positive, 213 negative
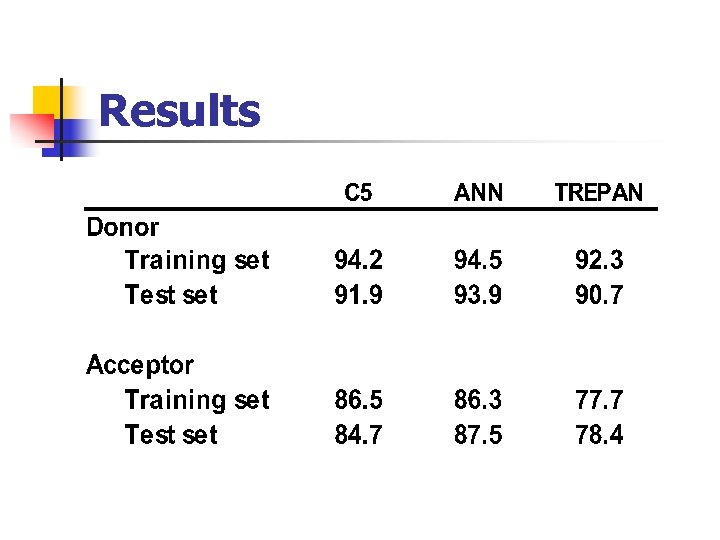
Results
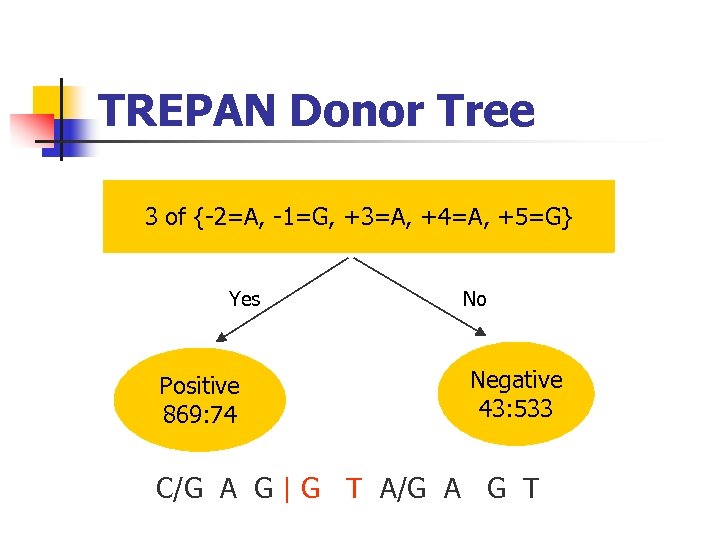
TREPAN Donor Tree 3 of {-2=A, -1=G, +3=A, +4=A, +5=G} Yes Positive 869: 74 No Negative 43: 533 C/G A G | G T A/G A G T
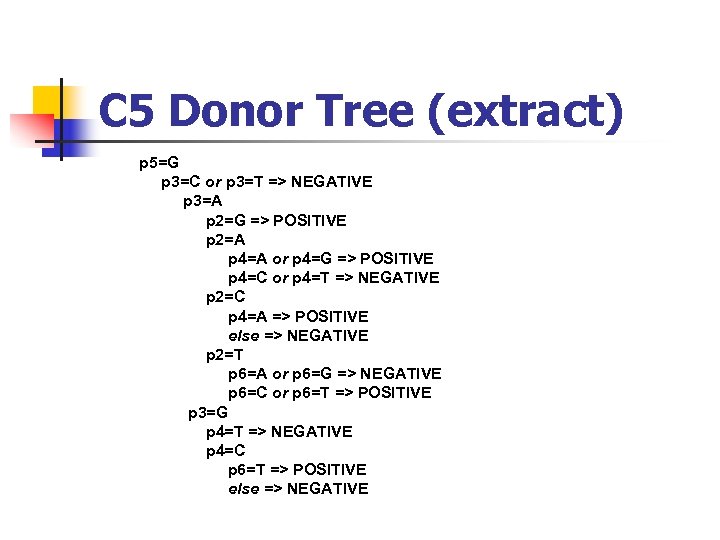
C 5 Donor Tree (extract) p 5=G p 3=C or p 3=T => NEGATIVE p 3=A p 2=G => POSITIVE p 2=A p 4=A or p 4=G => POSITIVE p 4=C or p 4=T => NEGATIVE p 2=C p 4=A => POSITIVE else => NEGATIVE p 2=T p 6=A or p 6=G => NEGATIVE p 6=C or p 6=T => POSITIVE p 3=G p 4=T => NEGATIVE p 4=C p 6=T => POSITIVE else => NEGATIVE
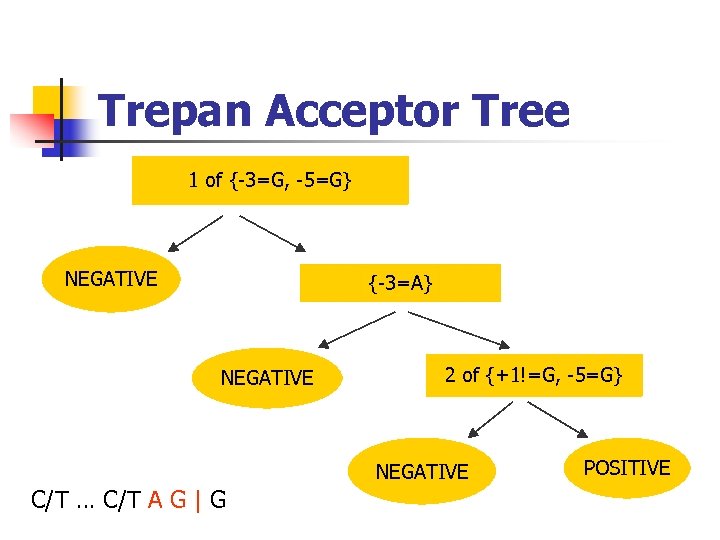
Trepan Acceptor Tree 1 of {-3=G, -5=G} NEGATIVE {-3=A} NEGATIVE 2 of {+1!=G, -5=G} NEGATIVE C/T … C/T A G | G POSITIVE
Application to Chemoinformatics 1. 2. 3. Learning general rules Conformational Analysis QSAR dataset
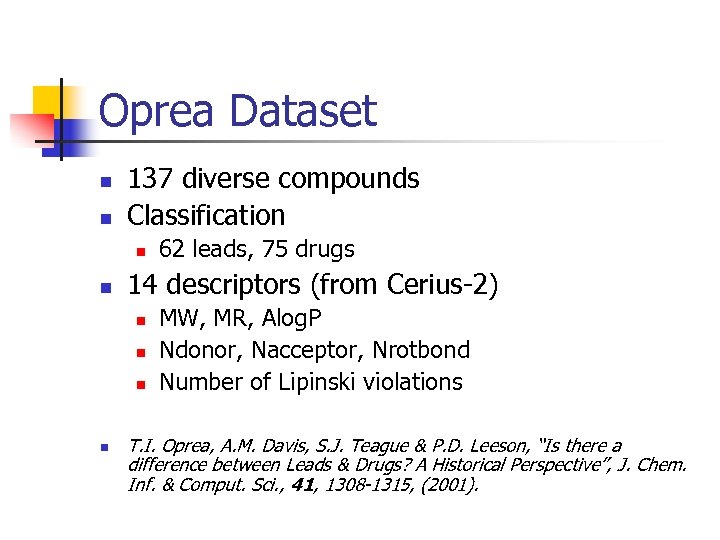
Oprea Dataset 137 diverse compounds Classification 14 descriptors (from Cerius-2) 62 leads, 75 drugs MW, MR, Alog. P Ndonor, Nacceptor, Nrotbond Number of Lipinski violations T. I. Oprea, A. M. Davis, S. J. Teague & P. D. Leeson, “Is there a difference between Leads & Drugs? A Historical Perspective”, J. Chem. Inf. & Comput. Sci. , 41, 1308 -1315, (2001).
![C 5 tree MW <= 380 [ Mode: lead ] Rule of 5 Violations C 5 tree MW <= 380 [ Mode: lead ] Rule of 5 Violations](https://present5.com/presentation/39904eb5a8510c001df6abfa091bbf23/image-17.jpg)
C 5 tree MW <= 380 [ Mode: lead ] Rule of 5 Violations = 0 [ Mode: lead ] Hbond acceptor <= 2 [ Mode: lead ] => lead Hbond acceptor > 2 [ Mode: drug ] => drug Rule of 5 Violations > 0 [ Mode: lead ] => lead MW > 380 [ Mode: drug ] => drug
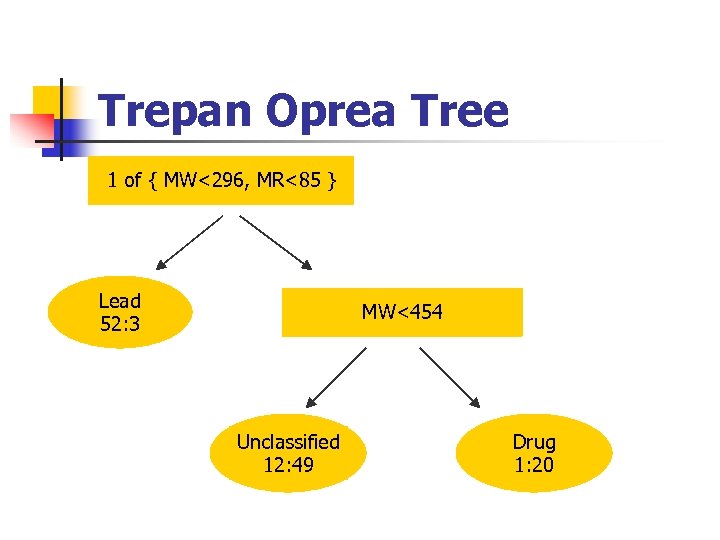
Trepan Oprea Tree 1 of { MW<296, MR<85 } Lead 52: 3 MW<454 Unclassified 12: 49 Drug 1: 20
Conformational Analysis 300 conformations from 5 ns MD simulation of rosiglitazone Classified by length of long axis into Extended – distance > 10 A Folded – distance < 10 A 8 torsion angles In house data.
Rosiglitazone Agonist of PPAR gamma Nuclear Receptor Regulates HDL/LDL and triglycerides Active ingredient of Avandia for Type II Diabetes
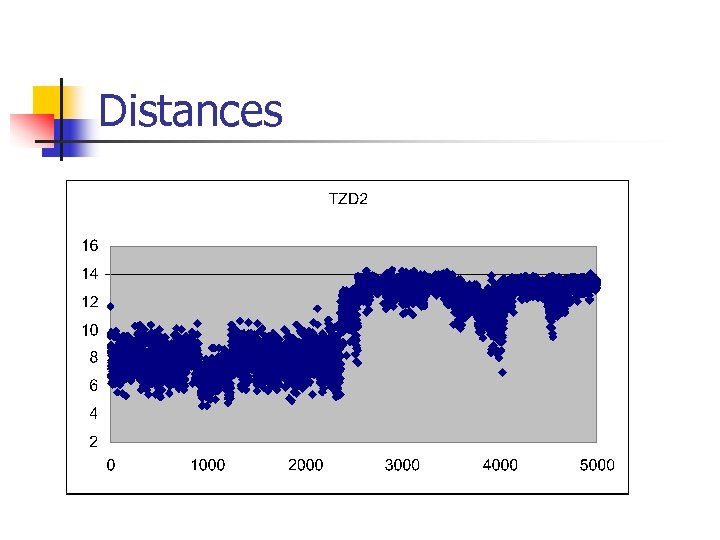
Distances
![C 5 tree T 5 <= 269 [ Mode: extended ] T 5 <= C 5 tree T 5 <= 269 [ Mode: extended ] T 5 <=](https://present5.com/presentation/39904eb5a8510c001df6abfa091bbf23/image-22.jpg)
C 5 tree T 5 <= 269 [ Mode: extended ] T 5 <= 52 [ Mode: extended ] T 7 <= 185 [ Mode: extended ] => extended T 7 > 185 [ Mode: folded ] T 6 <= 75 [ Mode: folded ] => folded T 6 > 75 [ Mode: extended ] T 5 <= 41 [ Mode: folded ] T 8 <= 249 [ Mode: folded ] => folded T 8 > 249 [ Mode: extended ] => extended T 5 > 41 [ Mode: extended ] => extended T 5 > 52 [ Mode: extended ] T 6 <= 73 [ Mode: extended ] T 8 <= 242 [ Mode: extended ] T 5 <= 7 [ Mode: extended ] T 8 <= 22 [ Mode: extended ] => extended T 8 > 22 [ Mode: folded ] => folded T 5 > 7 [ Mode: extended ] => extended T 8 > 242 [ Mode: extended ] => extended T 6 > 73 [ Mode: extended ] => extended T 5 > 269 [ Mode: folded ] => folded
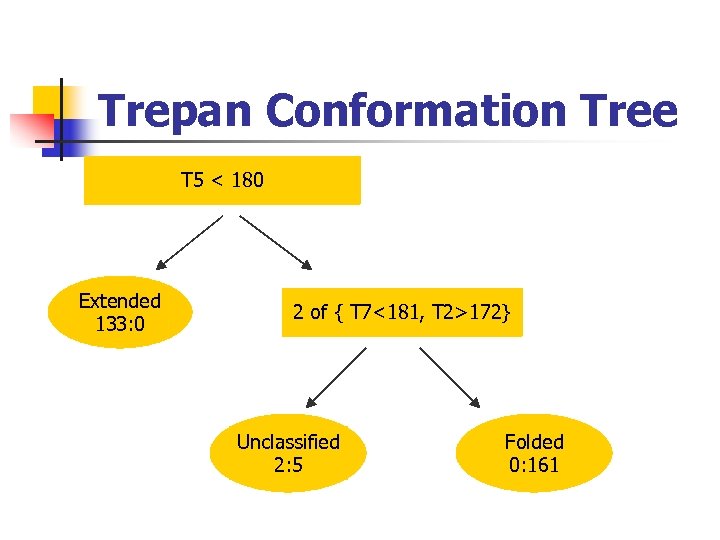
Trepan Conformation Tree T 5 < 180 Extended 133: 0 2 of { T 7<181, T 2>172} Unclassified 2: 5 Folded 0: 161
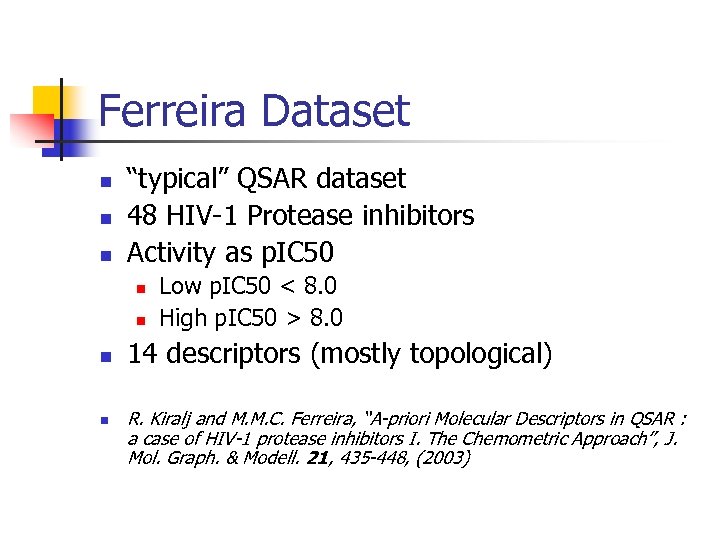
Ferreira Dataset “typical” QSAR dataset 48 HIV-1 Protease inhibitors Activity as p. IC 50 Low p. IC 50 < 8. 0 High p. IC 50 > 8. 0 14 descriptors (mostly topological) R. Kiralj and M. M. C. Ferreira, “A-priori Molecular Descriptors in QSAR : a case of HIV-1 protease inhibitors I. The Chemometric Approach”, J. Mol. Graph. & Modell. 21, 435 -448, (2003)

Original Results PLS model Activity determined by X 9, X 11, X 10, X 13 R 2 = 0. 91, Q 2=0. 85, Ncomps=3
![C 5 tree X 11 <= 2. 5 [ Mode: low ] X 13 C 5 tree X 11 <= 2. 5 [ Mode: low ] X 13](https://present5.com/presentation/39904eb5a8510c001df6abfa091bbf23/image-26.jpg)
C 5 tree X 11 <= 2. 5 [ Mode: low ] X 13 <= 16. 7 [ Mode: low ] => low X 13 > 16. 7 [ Mode: high ] => high X 11 > 2. 5 [ Mode: high ] => high
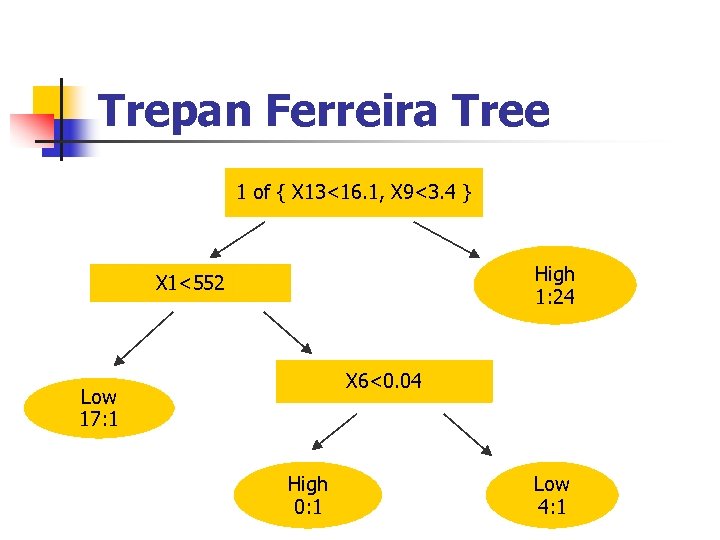
Trepan Ferreira Tree 1 of { X 13<16. 1, X 9<3. 4 } High 1: 24 X 1<552 X 6<0. 04 Low 17: 1 High 0: 1 Low 4: 1
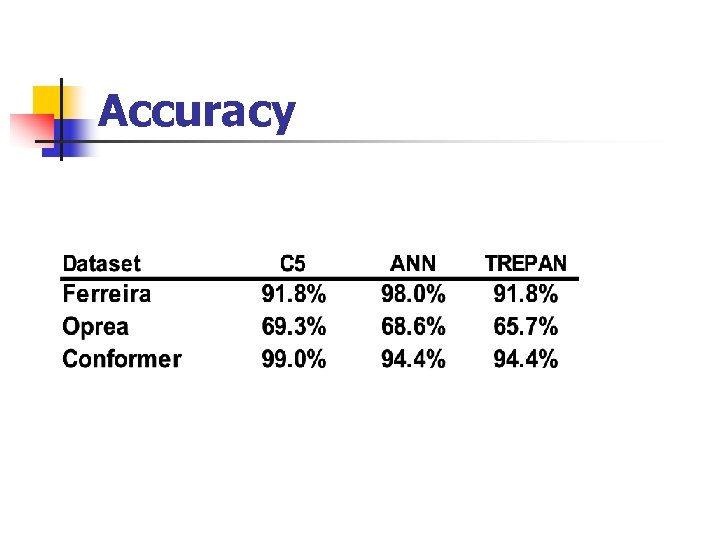
Accuracy

Conclusions Reasonable Accuracy Comprehensible Rules

Acknowledgements David Whitley. Tony Browne. Martyn Ford. BBSRC grant reference BIO/12005.
39904eb5a8510c001df6abfa091bbf23.ppt