c2530c63163b244d8a3a52d94d5087d7.ppt
- Количество слайдов: 35
Rule-based Knowledge Aggregation for Large-Scale Protein Sequence Analysis of Influenza A Viruses Sixth International Conference on Bioinformatics (In. Co. B 2007) Hong Kong, 28 th August 2007 Olivo Miotto Institute of Systems Science and Yong Loo Lin School of Medicine, National University of Singapore Tan Tin Wee Vladimir Brusic Yong Loo Lin School of Medicine National University of Singapore Cancer Vaccine Center Dana-Farber Cancer Inst.

Outline Knowledge Aggregation in large-scale analysis Semantic Technologies for Knowledge Aggregation Task: Annotating the Influenza Dataset XML-based structural rules Rule-based knowledge restructuring Discussion and Conclusions Page 2
Outline 1 Knowledge Aggregation in large-scale analysis Semantic Technologies for Knowledge Aggregation Task: Annotating the Influenza Dataset XML-based structural rules Rule-based knowledge restructuring Discussion and Conclusions Page 3
da ta Cu se rr is t p ent m rep ly, an a ua rat l io n Knowledge Aggregation: Scaling up Bioinformatics Bioinformatic Analysis is current limited in scope l l Usually single domain (single aspect) Mostly small datasets (single genes, or few sequences) "Horizontal" scalability: connecting domains l l l Multiple database sources, diversely purposed data Systemic and semantic heterogeneity Discovery by relationship analysis "Vertical" scalability: analyzing large datasets l l l Many thousands of records Diversity of geography, tissue types, host, etc. Discovery by comparative analysis Page 4
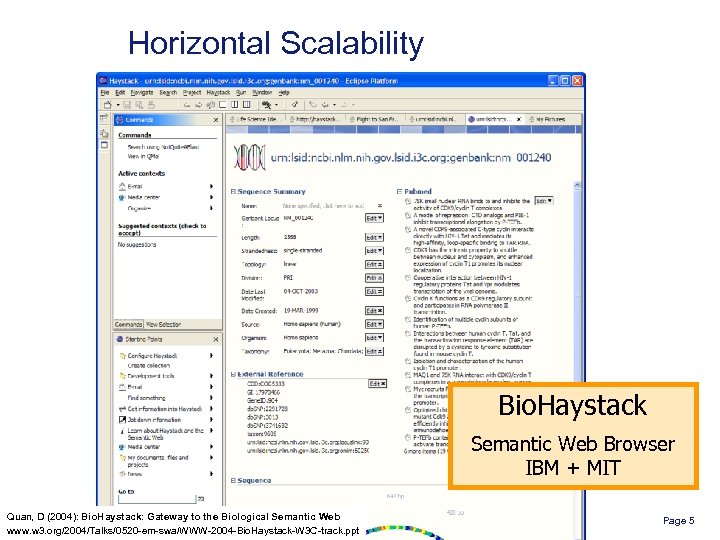
Horizontal Scalability Bio. Haystack Semantic Web Browser IBM + MIT Quan, D (2004): Bio. Haystack: Gateway to the Biological Semantic Web www. w 3. org/2004/Talks/0520 -em-swa/WWW-2004 -Bio. Haystack-W 3 C-track. ppt Page 5
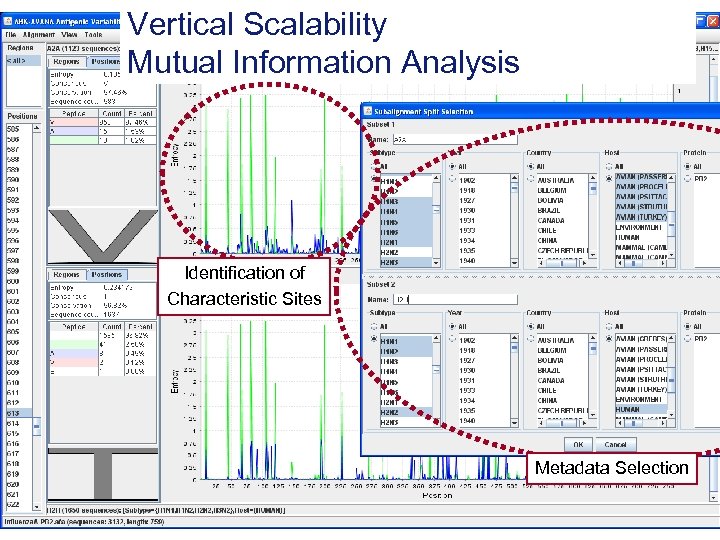
Vertical Scalability Mutual Information Analysis Identification of Characteristic Sites Metadata Selection Page 6
Obstacles to Scalability Heterogeneity of Biological Databases Systemic: access to data in different databases Syntactic: data formats, use of free text Structural: different table structures in different databases Semantic: data with different meaning and intent Semantic Heterogeneity is particularly insidious Data is rarely used in the way it was originally intended Low level of end-use technical expertise Biologists, not computer scientists Excel spreadsheets, Web page “scraping” Does not scale up Page 7
Outline Knowledge Aggregation in large-scale analysis 2 Semantic Technologies for Knowledge Aggregation Task: Annotating the Influenza Dataset XML-based structural rules Rule-based knowledge restructuring Discussion and Conclusions Page 8

Knowledge Aggregation: Technology requirements To enable large-scale Knowledge Aggregation we need a technology platform with l l Structural independence Structural adaptability To support biological researchers we need a technology platform with l l l Limited infrastructure needs Intutitiveness Easy interchange and transformation Best current candidate: Semantic Technologies Page 9
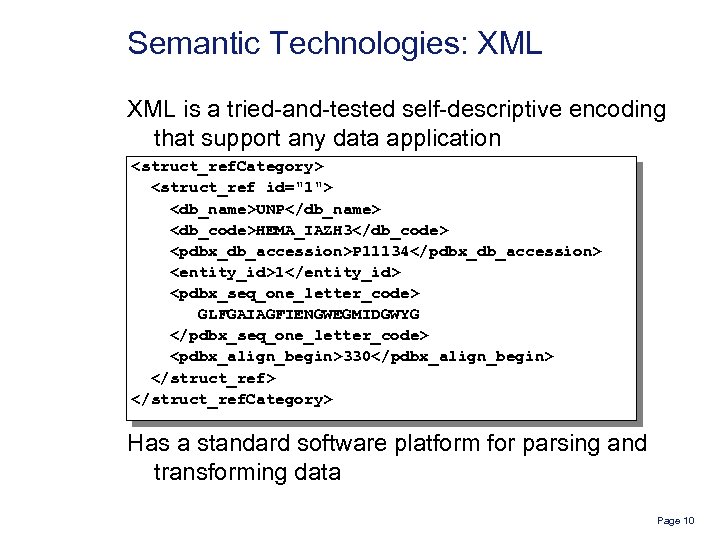
Semantic Technologies: XML is a tried-and-tested self-descriptive encoding that support any data application <struct_ref. Category> <struct_ref id="1"> <db_name>UNP</db_name> <db_code>HEMA_IAZH 3</db_code> <pdbx_db_accession>P 11134</pdbx_db_accession> <entity_id>1</entity_id> <pdbx_seq_one_letter_code> GLFGAIAGFIENGWEGMIDGWYG </pdbx_seq_one_letter_code> <pdbx_align_begin>330</pdbx_align_begin> </struct_ref. Category> Has a standard software platform for parsing and transforming data Page 10
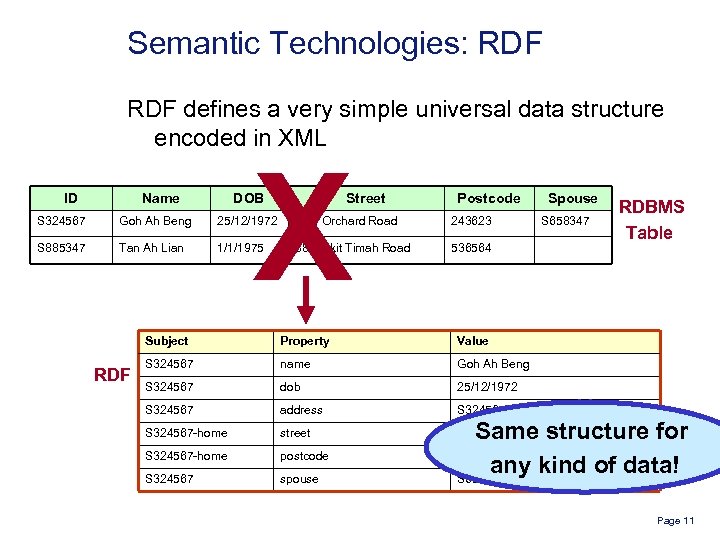
Semantic Technologies: RDF defines a very simple universal data structure encoded in XML ID Name X DOB Street Postcode S 324567 Goh Ah Beng 25/12/1972 127, Orchard Road 243623 S 885347 Tan Ah Lian 1/1/1975 88, Bukit Timah Road Spouse S 658347 536564 Subject RDF Property Value S 324567 name Goh Ah Beng S 324567 dob 25/12/1972 S 324567 address S 324567 -home street 127, Orchard Road S 324567 -home postcode S 324567 spouse RDBMS Table Same structure for 243623 any kind of data! S 658347 Page 11

Semantic Technologies: Ontologies: vocabularies of concepts and properties that describe a field of knowledge OWL technology allows user to define ontologies Shared ontologies allow interchange of data Ontologies support REASONING by means of programs that l l l Read RDF data, encoded using an ontology Apply rules that relate to the described properties Generate new knowledge from these rules Page 12
Outline Knowledge Aggregation in large-scale analysis Semantic Technologies for Knowledge Aggregation 3 Task: Annotating the Influenza Dataset XML-based structural rules Rule-based knowledge restructuring Discussion and Conclusions Page 13
Study goals Analyze all influenza protein sequences available l l Gen. Bank + Gen. Pept = 92, 343 documents Final dataset comprises 40, 169 unique sequences Various types of analysis, e. g. l l Identify amino acid mutations sites that characterize human-transmissible strains Compare the diversity of viral sequences over different periods of time and geographical areas Several Metadata fields required Protein name Host Subtype Country Isolate Year Manual Curation is not an Option! Page 14

Inconsistencies in Gen. Bank records Not so Good Pretty Bad Good Page 15

Experimental Approach 1. Retrieve all influenza A records from Gen. Bank and Genpept in XML format, using ABK platform Miotto O, Tan TW, Brusic V (2005) LNCS 3578, 398 -405. 2. Use XML structural rules to extract, merge and reconcile the metadata from the records 3. Use RDF encoding and an Ontology to encode and structure the resulting metadata 4. Use a Reasoner with Semantic Rules to restructure the metadata, and make inferences that improve the consistency Page 16

Outline Knowledge Aggregation in large-scale analysis Semantic Technologies for Knowledge Aggregation Task: Annotating the Influenza Dataset 4 XML-based structural rules Rule-based knowledge restructuring Discussion and Conclusions Page 17

Leveraging on XML offers great advantages for extracting heterogeneous metadata l l l Wide availability Popular encoding for source databases Standard processing software Independence from source schemas Query Language (XPath) Some disadvantages l l Almost unreadable by humans Interpretation of semantics requires understanding the schema Page 18
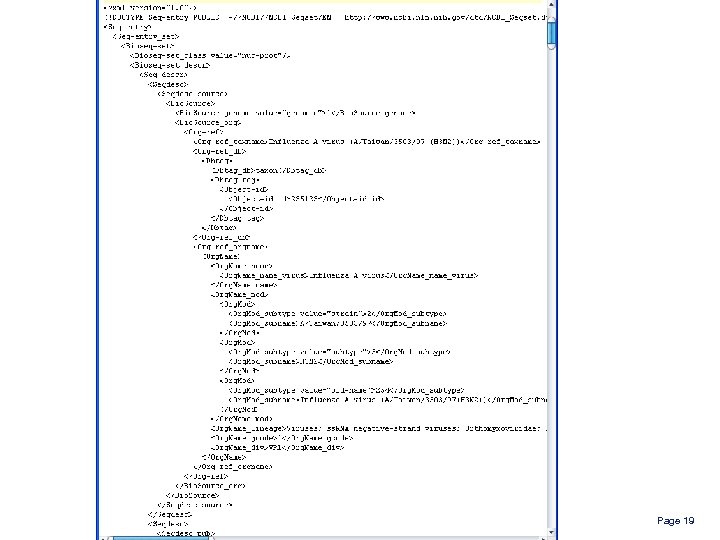
Page 19
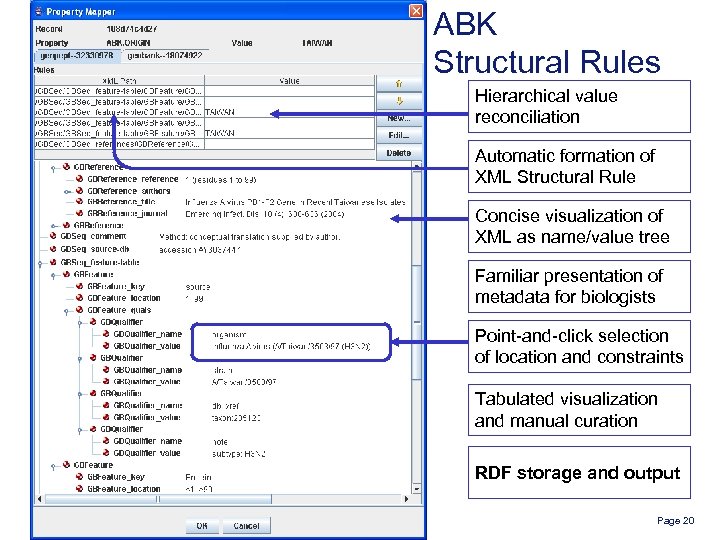
ABK Structural Rules Hierarchical value reconciliation Automatic formation of XML Structural Rule Concise visualization of XML as name/value tree Familiar presentation of metadata for biologists Point-and-click selection of location and constraints Tabulated visualization and manual curation RDF storage and output Page 20
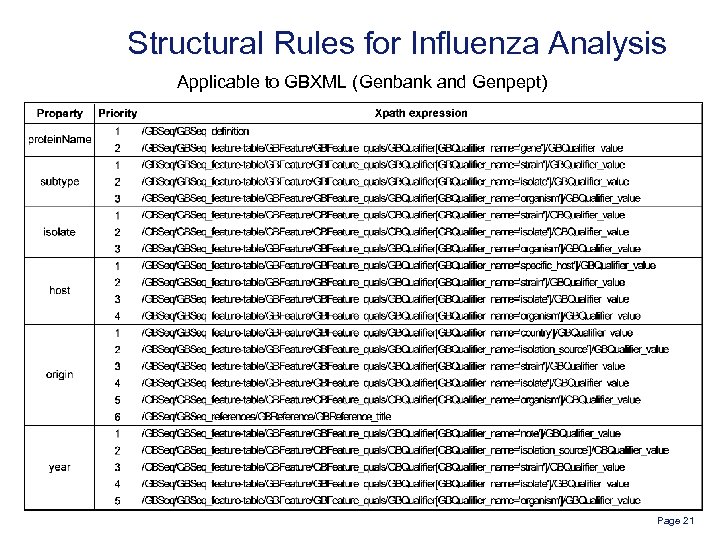
Structural Rules for Influenza Analysis Applicable to GBXML (Genbank and Genpept) Page 21
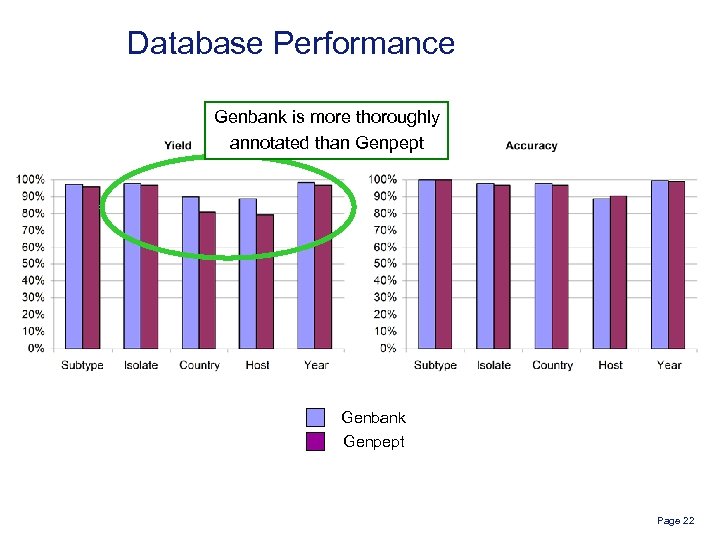
Database Performance Genbank is more thoroughly annotated than Genpept Genbank Genpept Page 22
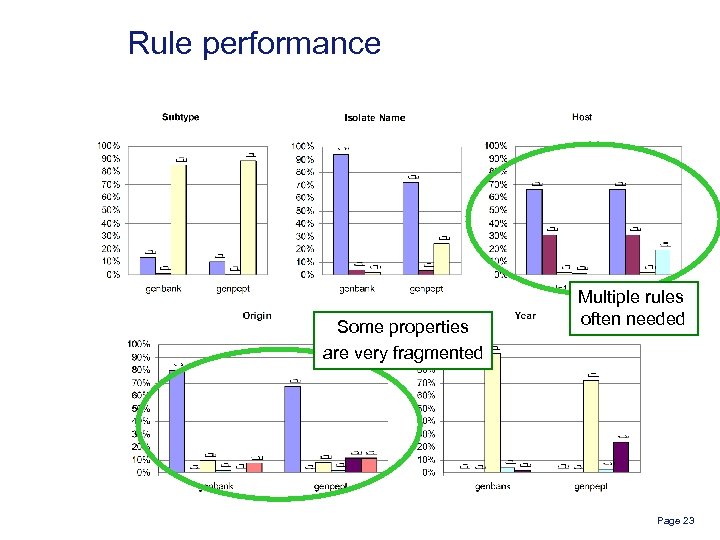
Rule performance Some properties are very fragmented Multiple rules often needed Page 23
Outline Knowledge Aggregation in large-scale analysis Semantic Technologies for Knowledge Aggregation Task: Annotating the Influenza Dataset XML-based structural rules 5 Rule-based knowledge restructuring Discussion and Conclusions Page 24
Semantic Metadata Restructuring Semantic Structure Gap Genbank semantics represents individual sequences A single isolate can comprise multiple sequences -> Sequences from same isolate can present metadata discrepancies Semantic Restructuring Restructure metadata to relate sequences from the same isolate Implemented using Jena 2 (http: //jena. sourceforge. net/) Native Jena rule-based reasoner Jena OWL reasoner validates inferences against ontology Page 25
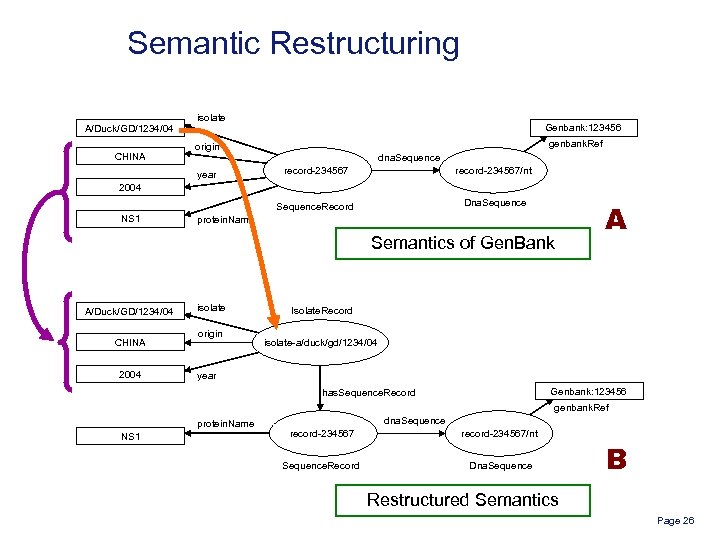
Semantic Restructuring A/Duck/GD/1234/04 CHINA isolate Genbank: 123456 genbank. Ref origin dna. Sequence record-234567/nt Sequence. Record year record-234567 Dna. Sequence 2004 NS 1 protein. Name Semantics of Gen. Bank A/Duck/GD/1234/04 CHINA 2004 isolate origin A Isolate. Record isolate-a/duck/gd/1234/04 year Genbank: 123456 has. Sequence. Record genbank. Ref protein. Name NS 1 dna. Sequence record-234567/nt Sequence. Record Dna. Sequence B Restructured Semantics Page 26

Restructuring Rules [rule 1: (? rec rdf: type vg: Sequence. Record) (? rec vg: isolate ? isolate. Id) normalize. Isolate(? isolate. Id, ? n. Iso. Id) uri. Concat('urn: abk: isolate: ', ? n. Iso. Id, ? isolate. Uri) -> (? isolate. Uri rdf: type vg: Isolate. Record) (? isolate. Uri vg: has. Sequence. Record ? rec) ] [rule 2: (? isolate. Uri vg: has. Sequence. Record ? rec) (? rec ? prop ? value) one. Of(? prop, vg: isolate, vg: virus. Subtype, vg: year, vg: country, vg: host. Organism) -> (? isolate. Uri ? prop ? value) ] Page 27
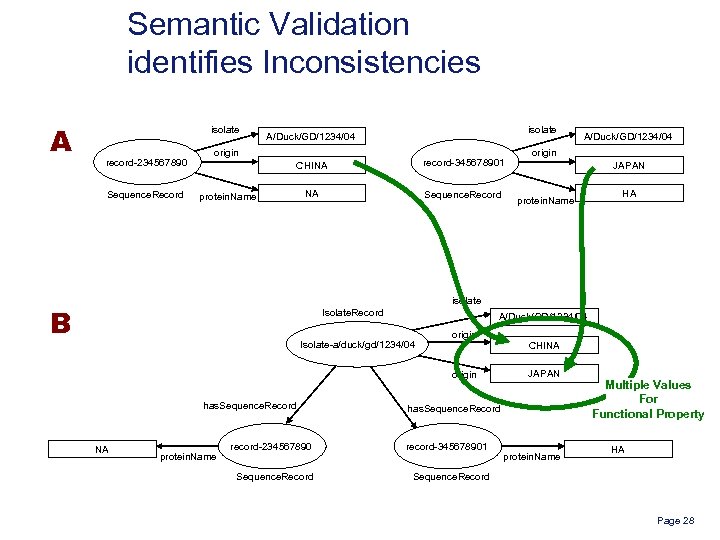
Semantic Validation identifies Inconsistencies A isolate record-234567890 Sequence. Record isolate A/Duck/GD/1234/04 origin CHINA record-345678901 NA Sequence. Record protein. Name A/Duck/GD/1234/04 origin JAPAN protein. Name HA isolate B Isolate. Record A/Duck/GD/1234/04 Isolate-a/duck/gd/1234/04 origin has. Sequence. Record NA protein. Name CHINA JAPAN has. Sequence. Record record-234567890 record-345678901 Sequence. Record Multiple Values For Functional Property Sequence. Record protein. Name HA Page 28
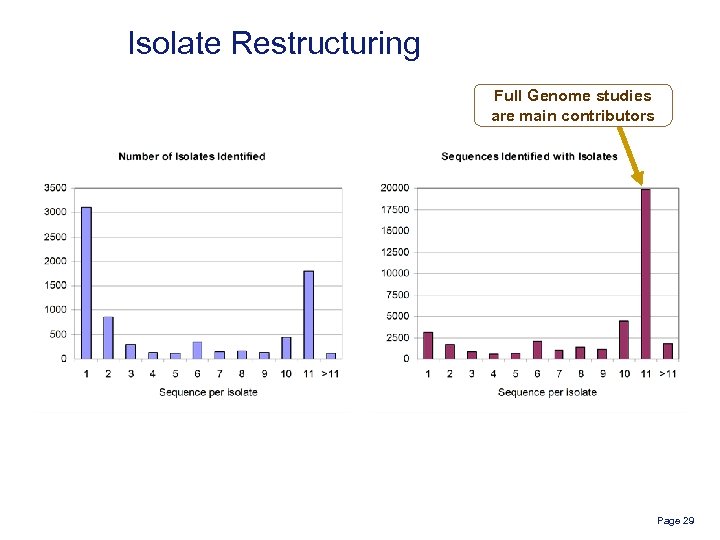
Isolate Restructuring Full Genome studies are main contributors Page 29
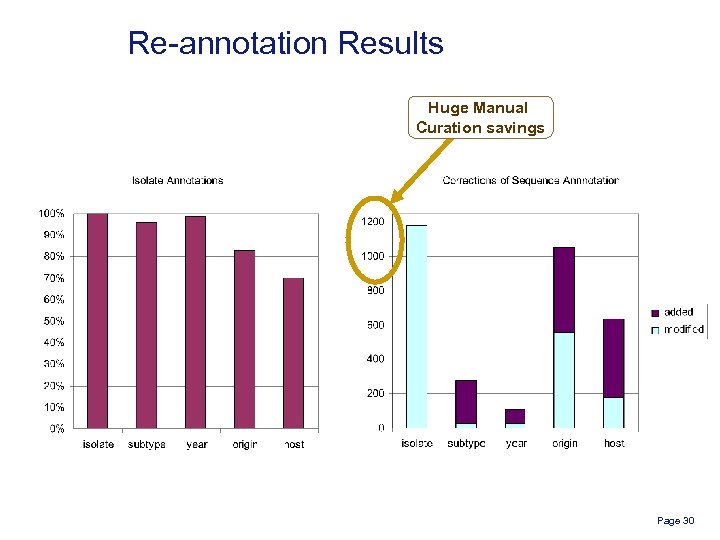
Re-annotation Results Huge Manual Curation savings Page 30
Outline Knowledge Aggregation in large-scale analysis Semantic Technologies for Knowledge Aggregation Task: Annotating the Influenza Dataset XML-based structural rules Rule-based knowledge restructuring 6 Discussion and Conclusions Page 31
Discussion - 1 Large-scale metadata recovery from public databases is difficult even for simple requirements Relatively simple approaches such as structural rules can do most of the tedious work l Accuracy can be further improved with machine learning Semantic inferences can improve data quality l Significant impact on manual curation task Rules have more potential for intuitive end-user GUI than programming l cf. email rules, firewall rules Page 32
Discussion - 2 Semantic Technologies are suitable for bioinformatics metadata management today l l Limited infrastructure requirements Flexibility and extensibility of ontologies (Open World) Enormous potential for analysis tool integration l Build tools that are "semantically agnostic" Reasoning currently computationally expensive l l l Our simple reasoning tasks exceeded the power of a current desktop when applied to 10, 000's records Divide-and conquer strategies were effective, but require manual work, and are not always applicable Reasoning services and computing grid can help scalability, but only if easy to access Page 33

Acknowledgements and Thanks Institute of Systems Science, NUS Funding support for this conference Prof. J Thomas August, Johns Hopkins University AT Heiny, NUS Partial Grant Support: National Institute of Allergy and Infectious Diseases, NIH Grant No. 5 U 19 AI 56541, Contract No. HHSN 2662 -00400085 C Immuno. Grid Project EC Contract FP 6 -2004 -IST-4, No. 028069 Page 34
Metadata Extraction Ontology (fragment) Sequence Record Class Six Functional Properties Page 35
c2530c63163b244d8a3a52d94d5087d7.ppt