RNAseq-Mapping-Vivek.pptx
- Количество слайдов: 39
RNA-Seq Reference Mapping based Analysis Plant Bioinformatics Workshop 2012 Tuesday, January 24 Vivek Krishnakumar

Constructing transcripts from RNA -Seq data Haas & Zody, 2010 http: //www. nature. com/nbt/journal/v 28/n 5/fig_tab/nbt 0510 -421_F 1. html
Benefits • Alignment to genome ¡ Allow reads from unannotated loci, introns etc. to align to their correct locations ¡ Potential for new biological insights – Detection of novel gene loci – Detection of possible functional non-coding RNA • Alignment to transcriptome ¡ Computationally inexpensive ¡ Spliced (junction) reads map correctly and easily ¡ Pairing distance and junction reads may help distinguish individual isoforms (informative/unique regions of transcripts)
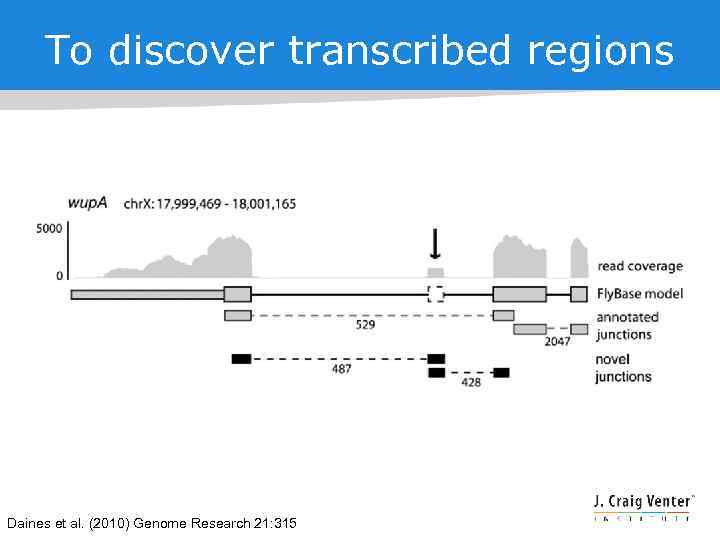
To discover transcribed regions Daines et al. (2010) Genome Research 21: 315
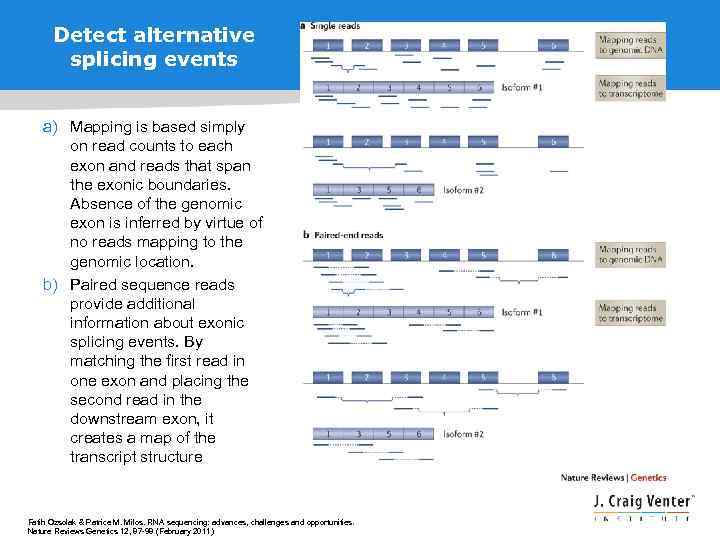
Detect alternative splicing events a) Mapping is based simply on read counts to each exon and reads that span the exonic boundaries. Absence of the genomic exon is inferred by virtue of no reads mapping to the genomic location. b) Paired sequence reads provide additional information about exonic splicing events. By matching the first read in one exon and placing the second read in the downstream exon, it creates a map of the transcript structure Fatih Ozsolak & Patrice M. Milos. RNA sequencing: advances, challenges and opportunities. Nature Reviews Genetics 12, 87 -98 (February 2011)
Drawbacks • Alignment to genome ¡ Computationally expensive ¡ It is never a good idea to simply align RNA-seq data to the genome. Need a spliced aligner • Alignment to transcriptome ¡ Reads deriving from non-genic regions may be ‘forcibly’ (and erroneously) aligned to genes – Incorrect gene expression values – False positive SNVs

Typical Analysis Workflow https: //www. msi. umn. edu/sites/default/files/RNA-Seq%20 Module 2%20 version%203. pdf

Quality Control • Recommended to use fastqc for the QC • Things to check for: ¡ Base calling reliability – ¡ Phred score >= 20 (per base & per sequence) Contaminations Overrepresented sequences – GC content – ¡ Synchronization of paired reads (make sure the reads are in proper order

Quality Trimming • If there are reads falling below the quality cutoff, they can be removed in entirety or bad bases can be trimmed off • Can be done using – FASTX toolkit – http: //hannonlab. cshl. edu/fastx_toolkit/galax y. html – trimmomatic – http: //www. usadellab. org/cms/index. php? pa ge=trimmomatic • If adapters are present (review the fastqc result), trim them off using Cut. Adapt or trimmomatic

Aligning Reads Bowtie BWA SHRi. MP SOAP BFAST MUMmer. GPU ERANGE free, open-source ELAND (part of Illumina's secondary analysis pipeline, CASAVA) free, closed-source Novoalign (Novocraft) CLCbio Genomics Workbench commercial, closed-source

Mapping Reads Back • Hash Table (Lookup table) ¡ FAST, but requires perfect matches. [O(m n + N)] • Dynamic Programming (Smith Waterman) ¡ ¡ ¡ Indels Mathematically optimal solution Slow (most programs use Hash Mapping as a prefilter) [O(mn. N)] • Burrows-Wheeler Transform (BW Transform) ¡ ¡ ¡ FAST. [O(m + N)] (without mismatch/gap) Memory efficient. But for gaps/mismatches, it lacks sensitivity

Why Burrows-Wheeler? Approximately ½ byte per base ¡ ¡ As large as the original text, plus a few “extras” Can fit onto a standard computer with 2 GB of memory Linear-time search algorithm ¡ proportional to length of query for exact matches Thanks to Haibao for the Burrow’s Wheeler slides

Burrows-Wheeler Transform (BWT) • Algorithm used for data compression • Output is easier to compress as it groups similar symbols together

Burrows-Wheeler Transform BWT acaacg$ $acaacg$ac acaacg$aca caacg$acaa g$acaac gc$aaac

Burrows-Wheeler Transform BWT acaacg$ 0123456 6 2 0 3 1 4 5 $acaacg$ac acaacg$aca caacg$acaa g$acaac gc$aaac

Burrows-Wheeler Transform BWT acaacg$ $acaacg$ac acaacg$aca caacg$acaa g$acaac gc$aaac This matrix has a property called last first (LF) mapping. The i-th occurrence of character X in the last column corresponds to the same text character as the i-th occurrence of X in the first column.

Exact search

Exact search acaacg$ 0123456 6 2 0 3 1 4 5 $acaacg$ac acaacg$aca caacg$acaa g$acaac Query sequence aac found at position 2!!

Burrows-Wheeler • Store entire reference genome. • Align tag base by base from the end. • When tag is traversed, all active locations are reported. • If no match is found, then back up and try a substitution.

Which aligner to choose? • Paired end alignment improves sensitivity • Use of (trusted) base quality could improve alignment BWA and Bowtie are the fastest (~7 Gbp per CPU day) Li, H and Homer, N (2010) Briefings in Bioinformatics 11: 473

BWA Burrow Wheelers Aligner • http: //bio-bwa. sourceforge. net • Very fast aligner • Finds all gapped alignments given a set of scoring parameters • Implements 2 algorithms ¡ bwa-short: query <= 200 bp – ¡ Li H. and Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics, 25: 1754 -60. [PMID: 19451168] bwa-sw: query <= 100 kbp – Li H. and Durbin R. (2010) Fast and accurate long-read alignment with Burrows-Wheeler Transform. Bioinformatics, Epub. [PMID: 20080505]

Bowtie • http: //bowtie-bio. sourceforge. net • Can map 35 -base-pair reads to the human genome at a rate of 25 million reads per hour • Indexes the genome with a Burrows-Wheeler index • Langmead B. , Trapnell C. , Pop M. , and Salzberg S. L. (2009) “Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. ” Genome Biology 10: R 25 [PMID: 19261174] • Does not perform gapped alignments • Possibility that it may not find all alignments based on the specified parameters

Top. Hat Trapnell C et al. Bioinformatics 2009; 25: 1105 -1111

Top. Hat Limitations & Considerations • Two‐step approach. If a read can be mapped to the genome without splicing, it would not be evaluated for spliced mapping • Canonical junctions only ¡ Reads < 75 bp, “GT‐AG” introns ¡ Reads >=75 bp, “GT‐AG”, “GC‐AG” and “AT‐AC” introns • Optimized for Human and mouse genomes. Parameters needs to changed to be more species specific. e. g. intron size

Mate Inner Distance • • Top. Hat makes use of the mate inner distance information in several places: • finding splice sites and fusion break points. • when choosing the best candidate alignments for pairs If mate inner distance is unknown, map some of the reads using Bowtie or BWA and depending on mapped position, estimate the isize

Running Top. Hat • Manual: http: //tophat. cbcb. umd. edu/manual. html • Requires a bowtie index of the reference genome tophat [options]* <index_base> <reads 1_1[, . . . , reads. N_1]> [reads 1_2, . . . reads. N_2] -r/--mate-inner-dist <int> Expected (mean) inner distance between mate pairs. Default 50 bp. -g/--max-multihits <int> Allow up to this many alignments to the reference for a given read. Default is 20 for read mapping. --library-type Top. Hat will treat the reads as strand specific -i/--min-intron-length <int> Minimum intron length. Default is 70. -I/--max-intron-length <int> Maximum intron length. Default is 500000. • Output: ¡ ¡ BAM file containing the accepted hits BED file containing a list of junctions identified (novel or otherwise)

Cufflinks Trapnell, C. , et al Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology (2010)

Issues with read counts Gene level expression: • Reads mapped to multiple locations of the genome ¡ uniformly divide each multi‐mapped read • Count reads mapped to all exons vs shared exons of all isoforms Isoform level expression • Assignment of reads to different isoforms ¡ maximum likelihood estimate

Quantifying Expression RPKM: Reads Per Kilobase per Million Mapped reads RPKM = C / LN C: Number of mappable reads on a feature (eg. Transcript, exon, etc. ) L: Length of feature (in kb) N: Total number of mappable reads (in millions) Mortazavi A. , et al. Mapping and quantifying mammalian transcriptomes by RNA‐Seq Nature Methods (2008)

Quantifying Expression RPKM http: //jura. wi. mit. edu/bio/education/hot_topics/RNAseq/RNA_Seq. pdf

Quantifying Expression FPKM: Fragments Per Kilobase of transcript per Million fragments mapped • Analogous to RPKM but does not use read counts • The relative abundances of transcripts are described in terms of the expected fragments observed from an RNA‐Seq experiment ¡ ¡ Uses statistical model to estimate likelihood for the abundance of set of transcripts, given the read fragments Probabilities are multiplied to find overall likelihood of observing the fragments in an experiment Trapnell, C. et al Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology (2010)

Handling multi-mapped reads • Individual reads may map to multiple regions because of: ¡ Sequence repeats ¡ Homology • Uniformly divide the reads to all mapping positions ¡ Calculate initial abundance for all transcripts based on the uniform dividing scheme ¡ Re-estimate abundance, dividing each MM read probabilistically based on initial abundance of genes to which it maps

Correcting sequence bias • • Sequence-specific bias is introduced during the library preparation; challenges the assumption of uniform coverage Bias usually caused by primers used either in PCR or reverse transcription (eg: bias caused by random hexamer priming) Appears near the ends of the sequences Correct bias by “learning” what sequences are being selected for ¡ Generate an initial abundance estimation without using bias correction ¡ Weight reads by the expression level of the transcript from which they arise to avoid over-counting sequences that may be common ¡ Revisit each fragment and apply the abundance while “learning” the features in a window around the 5’ and 3’ end, building a model for each end ¡ Re-estimate abundance based on the model built above and generate new set of FPKMs that are less affected by sequence bias

Running Cufflinks • Manual: http: //cufflinks. cbcb. umd. edu/manual. html • Takes Top. Hat accepted hits BAM file as input cufflinks [options]* <aligned_reads. (sam/bam)> -b/--frag-bias-correct <genome. fa> Providing Cufflinks with a multifasta file via this option instructs it to run our new bias detection and correction algorithm which can significantly improve accuracy of transcript abundance estimates -u/--multi-read-correct Tells Cufflinks to do an initial estimation procedure to more accurately weight reads mapping to multiple locations in the genome -I/--max-intron-length <int> The maximum intron length. Default is 300, 000. • Output: ¡ GTF file containing assembled transcripts ¡ Tracking files containing gene and isoform level expression values

Downstream Analyses • cuffcompare - analyze Cufflinks assembled transcripts • cuffmerge - merge together several Cufflinks assemblies • cuffdiff - find significant changes in transcript expression, splicing, and promoter use across replicates

Cuffcompare • Tool used to analyze the transcripts assembled by Cufflinks ¡ ¡ Compare against reference annotation Compare against other Cufflinks runs (time course experiments) • Input is the reference FASTA sequences and the GTF files to be compared • Output: ¡ stats – Accuracy of the transcripts in terms of Sensitivity (Sn) and Specificity (Sp) at gene, transcript, exon, intron and nucleotide level

Which approach is better? • Align-then-Assemble ¡ ¡ Preferred approach if reference genome is available Does not need abundant coverage for transcript discovery Able to discover less abundant isoforms Needs a reference genome • Assemble-then-Align ¡ ¡ ¡ The only approach possible when no reference genome is available Useful when transcripts are missing or incomplete in the reference genome Assembly of short reads is difficult Needs deep coverage for transcript discovery Unable to discover less abundant isoforms

Hybrid approach will probably perform the best Thanks to Suman Pakala (JCVI) for this slide

Questions?
RNAseq-Mapping-Vivek.pptx