41d6832b8c8fcffbc6dc87269e58a893.ppt
- Количество слайдов: 29
 Research Information Network Workshop Imperial College, London June 28 th 2006 Data Webs: new visions for research data on the Web Bio. Image. Web – integrating biological image data David Shotton Image Bio. Informatics Research Group Department of Zoology University of Oxford, UK e-mail: david. shotton @zoo. ox. ac. uk
Research Information Network Workshop Imperial College, London June 28 th 2006 Data Webs: new visions for research data on the Web Bio. Image. Web – integrating biological image data David Shotton Image Bio. Informatics Research Group Department of Zoology University of Oxford, UK e-mail: david. shotton @zoo. ox. ac. uk
 Outline n Integration of on-line data Ø The conventional approach Ø Heavyweight cross-database searching Ø The data web approach n Advantages of data webs n Scientific image data n The problem of locating research images n The special place of biological images n Bio. Image. Web – a data web for biological research images
Outline n Integration of on-line data Ø The conventional approach Ø Heavyweight cross-database searching Ø The data web approach n Advantages of data webs n Scientific image data n The problem of locating research images n The special place of biological images n Bio. Image. Web – a data web for biological research images
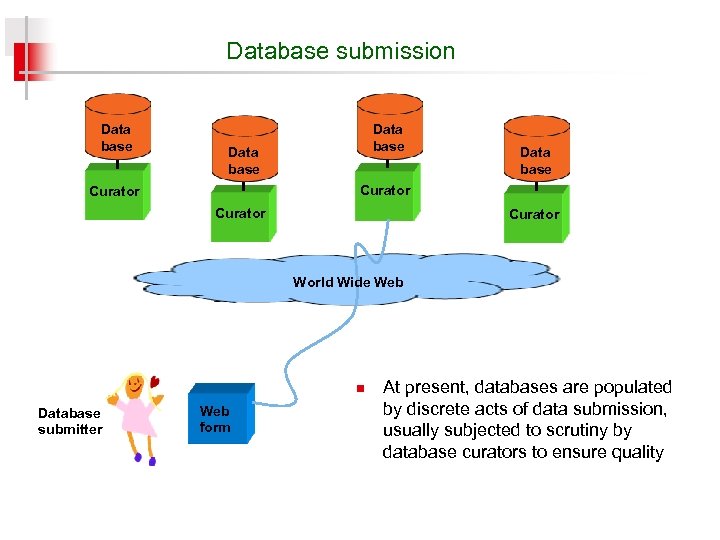 Database submission Data base Curator World Wide Web n Database submitter Web form At present, databases are populated by discrete acts of data submission, usually subjected to scrutiny by database curators to ensure quality
Database submission Data base Curator World Wide Web n Database submitter Web form At present, databases are populated by discrete acts of data submission, usually subjected to scrutiny by database curators to ensure quality
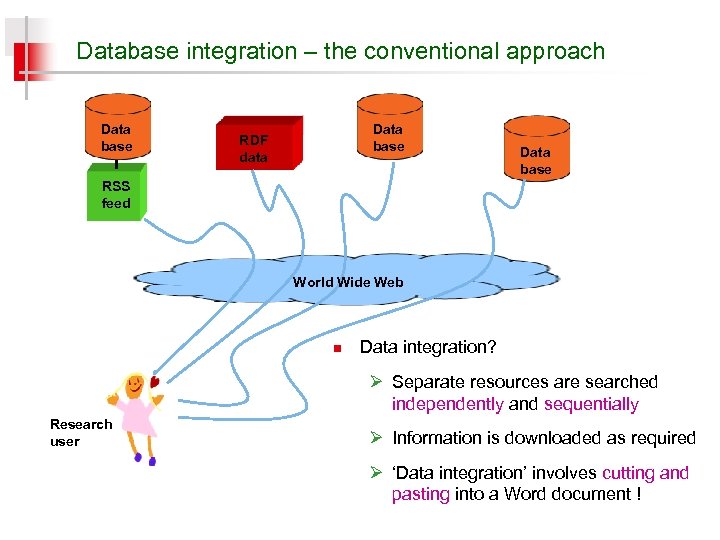 Database integration – the conventional approach Data base RDF data Data base RSS feed World Wide Web n Data integration? Ø Separate resources are searched independently and sequentially Research user Ø Information is downloaded as required Ø ‘Data integration’ involves cutting and pasting into a Word document !
Database integration – the conventional approach Data base RDF data Data base RSS feed World Wide Web n Data integration? Ø Separate resources are searched independently and sequentially Research user Ø Information is downloaded as required Ø ‘Data integration’ involves cutting and pasting into a Word document !
 Automated database integration – heavyweight approach n n Involves distributing SQL queries over geographically separate databases E. g. OGSA-DAI (Open Grid Services Architecture – Database Access and Integration) n Heavy investment from UK e-Science budget n Large development team n Heavyweight software solution Ø Involves user authorization and state maintenance n Has involved some interesting computer sciences, but has not been taken up for main-stream use by researchers!
Automated database integration – heavyweight approach n n Involves distributing SQL queries over geographically separate databases E. g. OGSA-DAI (Open Grid Services Architecture – Database Access and Integration) n Heavy investment from UK e-Science budget n Large development team n Heavyweight software solution Ø Involves user authorization and state maintenance n Has involved some interesting computer sciences, but has not been taken up for main-stream use by researchers!
 Automated data integration – the data web approach The data web concept is novel A data web is an alternative concept in digital information storage and integration n A step towards Berners-Lee’s vision of the Semantic Web as a ‘Web of Data’ Ø The creation of subject-specific data webs to meet defined research needs Ø This approach is particularly appropriate for data representing ‘particulars’ n n The data are NOT submitted to a central database, but are simply published by the data providers on their own Web servers Then, separately for each data web serving a particular knowledge domain, lightweight software tools are used to harvest, marshal and index metadata describing the distributed data into a central ontology-enabled data web registry
Automated data integration – the data web approach The data web concept is novel A data web is an alternative concept in digital information storage and integration n A step towards Berners-Lee’s vision of the Semantic Web as a ‘Web of Data’ Ø The creation of subject-specific data webs to meet defined research needs Ø This approach is particularly appropriate for data representing ‘particulars’ n n The data are NOT submitted to a central database, but are simply published by the data providers on their own Web servers Then, separately for each data web serving a particular knowledge domain, lightweight software tools are used to harvest, marshal and index metadata describing the distributed data into a central ontology-enabled data web registry
 Data web metadata harvesting n n n In the loosely coupled world of data webs, strict metadata conformity is not required To create a data web, you take what you can get and make the most of it ! All that is required of the data publisher is to make her existing metadata available on her server Ø in conformance to a minimalist core metadata schema Ø using the data web namespace n n These metadata are then harvested and integrated automatically The data web overcomes the problems caused by differences in data presentation, both syntactic and semantic, and makes collating selected information from multiple web sites possible for machines
Data web metadata harvesting n n n In the loosely coupled world of data webs, strict metadata conformity is not required To create a data web, you take what you can get and make the most of it ! All that is required of the data publisher is to make her existing metadata available on her server Ø in conformance to a minimalist core metadata schema Ø using the data web namespace n n These metadata are then harvested and integrated automatically The data web overcomes the problems caused by differences in data presentation, both syntactic and semantic, and makes collating selected information from multiple web sites possible for machines
 Data web metadata adaptation n n n We would ideally like all metadata to be presented as RDF However, most on-line data repositories and databases currently use XML or relational (tabular) data formats We do not see this as a major obstacle, since we have successfully used SPARQL with the open source tool D 2 RQ (http: //www. wiwiss. fuberlin. de/suhl/bizer/D 2 RQ/) to query relational databases in terms of an RDF data model, and we anticipate that similar tools will soon be available for querying XML data We thus believe that extant non-RDF data sources can be combined with ‘born RDF’ data, without the necessity for extensive reformatting of the non-RDF data Only minimal adaptation of existing metadata will be mandated, just sufficient to ensure consistency, truth maintenance and cross-referencing between heterogeneous repositories This adaptation will involve both straightforward syntactic transformations, and also conformity to an appropriate data web core ontology, to ensure semantic consistency for a minimal set of metadata terms between various repositories
Data web metadata adaptation n n n We would ideally like all metadata to be presented as RDF However, most on-line data repositories and databases currently use XML or relational (tabular) data formats We do not see this as a major obstacle, since we have successfully used SPARQL with the open source tool D 2 RQ (http: //www. wiwiss. fuberlin. de/suhl/bizer/D 2 RQ/) to query relational databases in terms of an RDF data model, and we anticipate that similar tools will soon be available for querying XML data We thus believe that extant non-RDF data sources can be combined with ‘born RDF’ data, without the necessity for extensive reformatting of the non-RDF data Only minimal adaptation of existing metadata will be mandated, just sufficient to ensure consistency, truth maintenance and cross-referencing between heterogeneous repositories This adaptation will involve both straightforward syntactic transformations, and also conformity to an appropriate data web core ontology, to ensure semantic consistency for a minimal set of metadata terms between various repositories
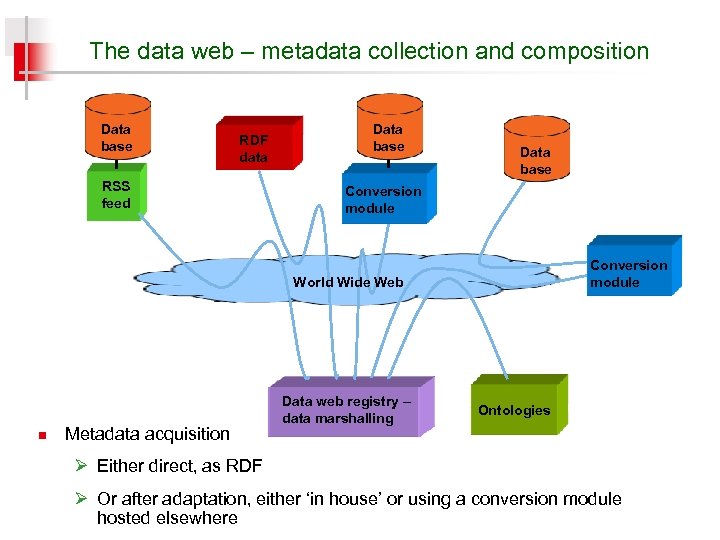 The data web – metadata collection and composition Data base RDF data RSS feed Data base Conversion module World Wide Web n Metadata acquisition Data web registry – data marshalling Ontologies Ø Either direct, as RDF Ø Or after adaptation, either ‘in house’ or using a conversion module hosted elsewhere
The data web – metadata collection and composition Data base RDF data RSS feed Data base Conversion module World Wide Web n Metadata acquisition Data web registry – data marshalling Ontologies Ø Either direct, as RDF Ø Or after adaptation, either ‘in house’ or using a conversion module hosted elsewhere
 Role of the central metadata registry n The data web registry acts first as a data marshal, gathering, ordering and integrating the metadata from across the web into a single searchable RDF graph Ø Remember: With RDF, integration comes for free! n n It then provides an integrated cross-searchable access point to all the data in the data web, with both human and programmatic access The data web registry adds value by providing interoperability and customizable search interfaces, with a rigorous semantic underpinning The primary data holders benefit by increased user traffic to their sites, while at the same time being able to maintain normal copyright and access control The primary metadata are never controlled by the data web registry, but are freely available on the Web for use by other presently unforeseen applications, including novel data mining, integration and analysis services
Role of the central metadata registry n The data web registry acts first as a data marshal, gathering, ordering and integrating the metadata from across the web into a single searchable RDF graph Ø Remember: With RDF, integration comes for free! n n It then provides an integrated cross-searchable access point to all the data in the data web, with both human and programmatic access The data web registry adds value by providing interoperability and customizable search interfaces, with a rigorous semantic underpinning The primary data holders benefit by increased user traffic to their sites, while at the same time being able to maintain normal copyright and access control The primary metadata are never controlled by the data web registry, but are freely available on the Web for use by other presently unforeseen applications, including novel data mining, integration and analysis services
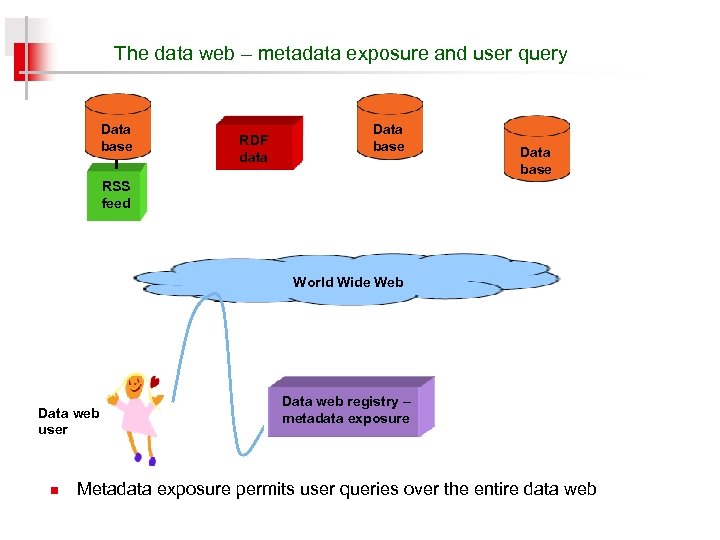 The data web – metadata exposure and user query Data base RDF data Data base RSS feed World Wide Web Data web user n Data web registry – metadata exposure Metadata exposure permits user queries over the entire data web
The data web – metadata exposure and user query Data base RDF data Data base RSS feed World Wide Web Data web user n Data web registry – metadata exposure Metadata exposure permits user queries over the entire data web
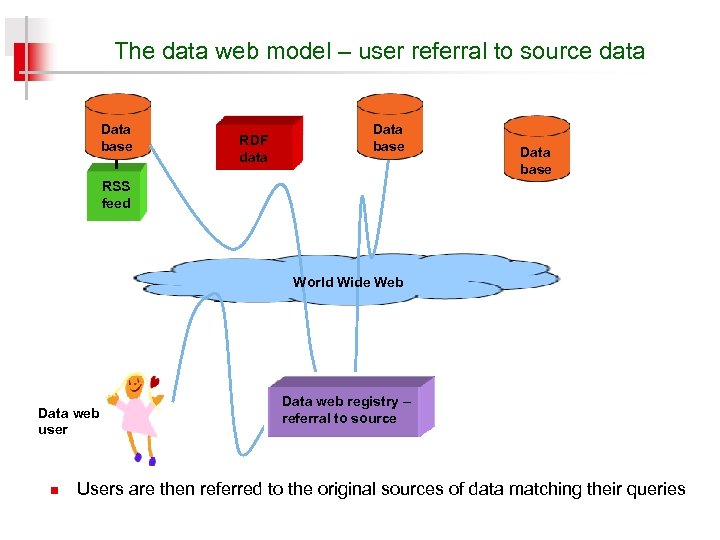 The data web model – user referral to source data Data base RDF data Data base RSS feed World Wide Web Data web user n Data web registry – referral to source Users are then referred to the original sources of data matching their queries
The data web model – user referral to source data Data base RDF data Data base RSS feed World Wide Web Data web user n Data web registry – referral to source Users are then referred to the original sources of data matching their queries
 Data web advantages n A data web of this type will have all the advantages of the World Wide Web itself: Ø distributed data Ø freedom, decentralization and low cost of publication Ø lack of centralized control Ø a "missing isn’t broken" Open World philosophy Ø built-in evolvability and scalability
Data web advantages n A data web of this type will have all the advantages of the World Wide Web itself: Ø distributed data Ø freedom, decentralization and low cost of publication Ø lack of centralized control Ø a "missing isn’t broken" Open World philosophy Ø built-in evolvability and scalability
 How does a data webs differ from Google? n A data web will provide for the selected data what Web search engines such as Google do for conventional Web pages, but with the following advantages: Ø It involves specific targeting to a particular knowledge domain, thus achieving a significantly higher signal-to-noise ratio Ø It provides integration of information with ontological underpinning, semantic coherence, and truth maintenance Ø It permits programmatic access, enabling further services to be built on top of one or more data webs
How does a data webs differ from Google? n A data web will provide for the selected data what Web search engines such as Google do for conventional Web pages, but with the following advantages: Ø It involves specific targeting to a particular knowledge domain, thus achieving a significantly higher signal-to-noise ratio Ø It provides integration of information with ontological underpinning, semantic coherence, and truth maintenance Ø It permits programmatic access, enabling further services to be built on top of one or more data webs
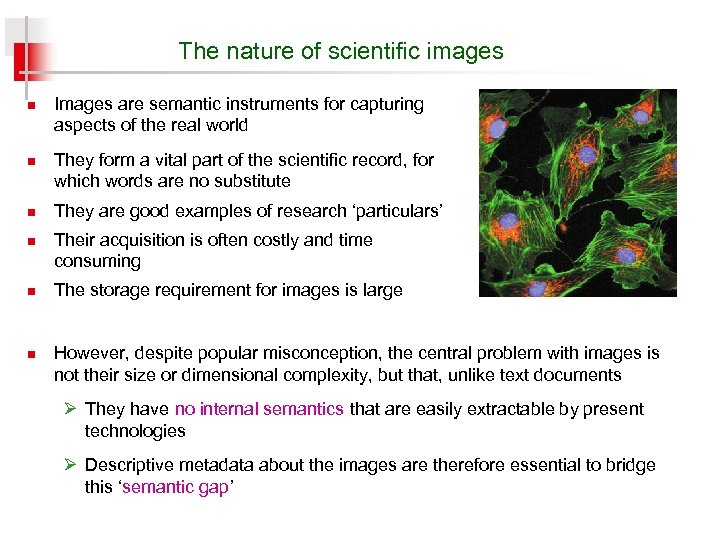 The nature of scientific images n n n Images are semantic instruments for capturing aspects of the real world They form a vital part of the scientific record, for which words are no substitute They are good examples of research ‘particulars’ Their acquisition is often costly and time consuming The storage requirement for images is large However, despite popular misconception, the central problem with images is not their size or dimensional complexity, but that, unlike text documents Ø They have no internal semantics that are easily extractable by present technologies Ø Descriptive metadata about the images are therefore essential to bridge this ‘semantic gap’
The nature of scientific images n n n Images are semantic instruments for capturing aspects of the real world They form a vital part of the scientific record, for which words are no substitute They are good examples of research ‘particulars’ Their acquisition is often costly and time consuming The storage requirement for images is large However, despite popular misconception, the central problem with images is not their size or dimensional complexity, but that, unlike text documents Ø They have no internal semantics that are easily extractable by present technologies Ø Descriptive metadata about the images are therefore essential to bridge this ‘semantic gap’
 The problem of finding images on the Web n n The value of on-line digital image information depends upon how easily it can be located and retrieved Without appropriately structured metadata, on-line digital image repositories become little more than meaningless and costly data graveyards n Even worse, only a small fraction of research images are ever published n With on-line journals, publication of multiple images has become easier n But such ‘supplementary data’ in on-line journals often Ø require subscription access Ø are not well annotated Ø are not cross-searchable n There is no general means at present of searching across different Web image repositories except using Google Images or an equivalent
The problem of finding images on the Web n n The value of on-line digital image information depends upon how easily it can be located and retrieved Without appropriately structured metadata, on-line digital image repositories become little more than meaningless and costly data graveyards n Even worse, only a small fraction of research images are ever published n With on-line journals, publication of multiple images has become easier n But such ‘supplementary data’ in on-line journals often Ø require subscription access Ø are not well annotated Ø are not cross-searchable n There is no general means at present of searching across different Web image repositories except using Google Images or an equivalent
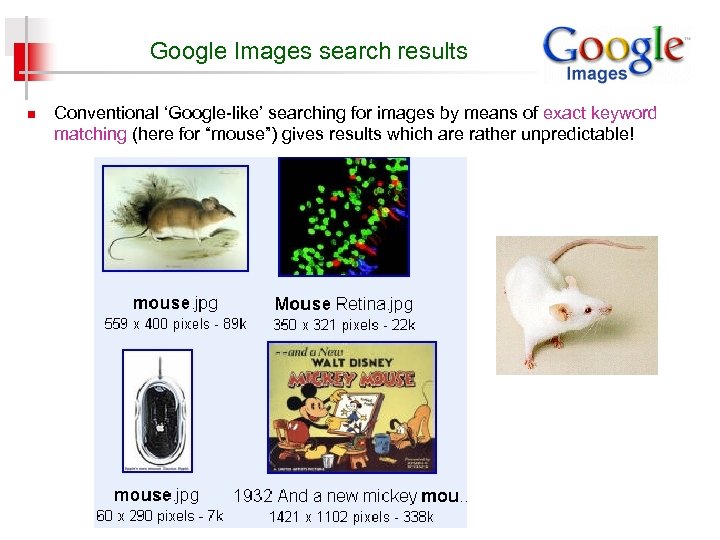 Google Images search results n Conventional ‘Google-like’ searching for images by means of exact keyword matching (here for “mouse”) gives results which are rather unpredictable!
Google Images search results n Conventional ‘Google-like’ searching for images by means of exact keyword matching (here for “mouse”) gives results which are rather unpredictable!
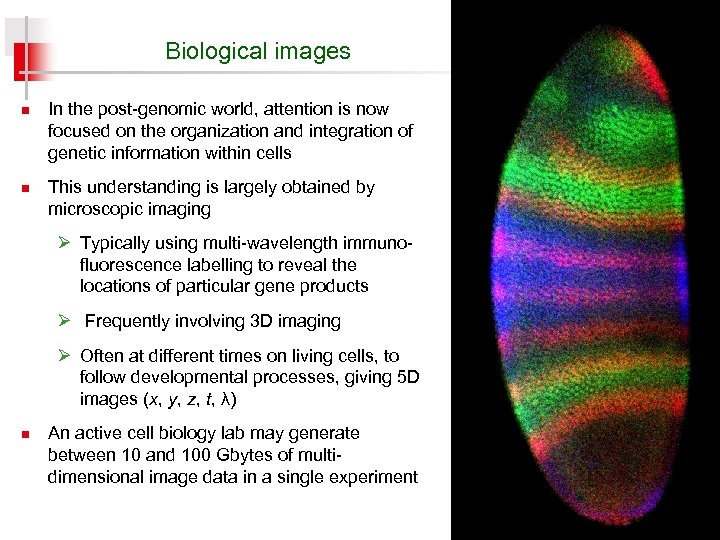 Biological images n n In the post-genomic world, attention is now focused on the organization and integration of genetic information within cells This understanding is largely obtained by microscopic imaging Ø Typically using multi-wavelength immunofluorescence labelling to reveal the locations of particular gene products Ø Frequently involving 3 D imaging Ø Often at different times on living cells, to follow developmental processes, giving 5 D images (x, y, z, t, λ) n An active cell biology lab may generate between 10 and 100 Gbytes of multidimensional image data in a single experiment
Biological images n n In the post-genomic world, attention is now focused on the organization and integration of genetic information within cells This understanding is largely obtained by microscopic imaging Ø Typically using multi-wavelength immunofluorescence labelling to reveal the locations of particular gene products Ø Frequently involving 3 D imaging Ø Often at different times on living cells, to follow developmental processes, giving 5 D images (x, y, z, t, λ) n An active cell biology lab may generate between 10 and 100 Gbytes of multidimensional image data in a single experiment
 A starting place for data webs - biological images n n n As particulars rather than universals, it is not appropriate to harvest all research images into a central database Neither would that solution scale well, since images are orders of magnitude larger than genomic sequences Furthermore, unlike biological sequences and molecular crystal structures, which carry their own internal semantics, images are not self-describing Thus, to be properly understood, a biological research image must either be accompanied by rich metadata, or be viewable in the context of a paper giving the full experimental description and interpretation relating to it Bio. Image. Web will be an exemplar data web, specific for published biological research images web What is it? A river delta? Broccoli? A lymphocyte cell?
A starting place for data webs - biological images n n n As particulars rather than universals, it is not appropriate to harvest all research images into a central database Neither would that solution scale well, since images are orders of magnitude larger than genomic sequences Furthermore, unlike biological sequences and molecular crystal structures, which carry their own internal semantics, images are not self-describing Thus, to be properly understood, a biological research image must either be accompanied by rich metadata, or be viewable in the context of a paper giving the full experimental description and interpretation relating to it Bio. Image. Web will be an exemplar data web, specific for published biological research images web What is it? A river delta? Broccoli? A lymphocyte cell?
 The Bio. Image. Web Consortium n Image Bio. Informatics Research Group, University of Oxford n Leading commercial publishers Ø Nature and Oxford University Press n Leading Open Access publishers Ø The Public Library of Science and Bio. Med Central n University institutional repositories Ø Universities of Cambridge, Imperial College, Oxford and Southampton n Other stakeholders Ø NHM, British Library, CCLRC, UKOLN, ILRT, Cross. Ref, and SPARC Europe n Professional biologists and academic biological image collections
The Bio. Image. Web Consortium n Image Bio. Informatics Research Group, University of Oxford n Leading commercial publishers Ø Nature and Oxford University Press n Leading Open Access publishers Ø The Public Library of Science and Bio. Med Central n University institutional repositories Ø Universities of Cambridge, Imperial College, Oxford and Southampton n Other stakeholders Ø NHM, British Library, CCLRC, UKOLN, ILRT, Cross. Ref, and SPARC Europe n Professional biologists and academic biological image collections
 Bio. Image. Web - aims and benefits n n n To integrate and make cross-searchable published biological research images currently held in isolated data silos To require minimum effort on the part of the image owners, who can use their existing relational databases, XML metadata schemas or RSS feeds To bring users to the image sites, while allowing publishers to retain access control to their own journals, and copyright holders to maintain copyright over their primary image data To enable publishers’ web sites to become more integral parts of the day-today information environment of biological researchers To permit owners of research image collections to publish their images in a manner that can easily integrate with other research image collections web
Bio. Image. Web - aims and benefits n n n To integrate and make cross-searchable published biological research images currently held in isolated data silos To require minimum effort on the part of the image owners, who can use their existing relational databases, XML metadata schemas or RSS feeds To bring users to the image sites, while allowing publishers to retain access control to their own journals, and copyright holders to maintain copyright over their primary image data To enable publishers’ web sites to become more integral parts of the day-today information environment of biological researchers To permit owners of research image collections to publish their images in a manner that can easily integrate with other research image collections web
 Bio. Image. Web – an (imperfect) real world analogy n n The local newspaper property section contains thumbnail images and basic metadata about houses for sale – equivalent to the Bio. Image. Web registry Users searching this central ‘registry’ pick out what they like, and then. . . Thumbnail image Minimal metadata
Bio. Image. Web – an (imperfect) real world analogy n n The local newspaper property section contains thumbnail images and basic metadata about houses for sale – equivalent to the Bio. Image. Web registry Users searching this central ‘registry’ pick out what they like, and then. . . Thumbnail image Minimal metadata
 . . . go round to the estate agent’s office for full details!
. . . go round to the estate agent’s office for full details!
 Bio. Image. Web – requirements n To develop a ‘Bio. Image. Core’ ontology using extended Dublin Core n To map data providers’ metadata schemas to Bio. Image. Core n n n To use agent technology to recognise sites conforming to the Bio. Image. Web namespace To develop server modules that automatically harvest providers’ metadata and image thumbnails as RDF To maintain the core Bio. Image. Web metadata marshalling registry To provide both user interfaces and programmatic access for searching across these integrated metadata To maintain links from the search results back to the original image data web
Bio. Image. Web – requirements n To develop a ‘Bio. Image. Core’ ontology using extended Dublin Core n To map data providers’ metadata schemas to Bio. Image. Core n n n To use agent technology to recognise sites conforming to the Bio. Image. Web namespace To develop server modules that automatically harvest providers’ metadata and image thumbnails as RDF To maintain the core Bio. Image. Web metadata marshalling registry To provide both user interfaces and programmatic access for searching across these integrated metadata To maintain links from the search results back to the original image data web
 Summary - the data web philosophy n Our ‘publish locally then integrate’ data web philosophy, which stands in contrast to the formal submission policies of central databases, is: Ø to harvest metadata describing heterogeneous data in a particular subject area, published on distributed Web sites Ø to aggregate, marshal and index them in a domain-specific data web registry that will provide both human and programmatic access Ø and to sustain links back to the data sources from which users can download or compare the original published data that meet their search criteria n n In principal, different data webs might access the same data sources for quite distinct purposes, for example accessing cellular images either to compare microscopy techniques or to study disease progression We believe that such a generic light-weight data web approach, involving a search across aggregated metadata, will ultimately prove to be a better and more scalable method than searching directly across multiple databases
Summary - the data web philosophy n Our ‘publish locally then integrate’ data web philosophy, which stands in contrast to the formal submission policies of central databases, is: Ø to harvest metadata describing heterogeneous data in a particular subject area, published on distributed Web sites Ø to aggregate, marshal and index them in a domain-specific data web registry that will provide both human and programmatic access Ø and to sustain links back to the data sources from which users can download or compare the original published data that meet their search criteria n n In principal, different data webs might access the same data sources for quite distinct purposes, for example accessing cellular images either to compare microscopy techniques or to study disease progression We believe that such a generic light-weight data web approach, involving a search across aggregated metadata, will ultimately prove to be a better and more scalable method than searching directly across multiple databases
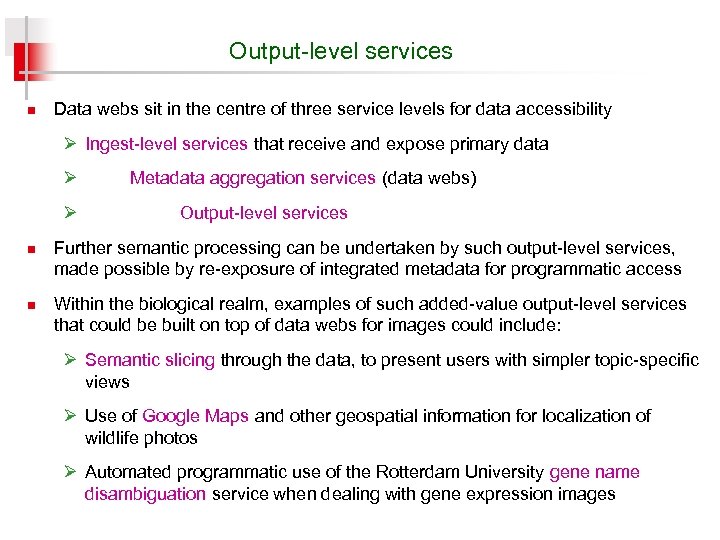 Output-level services n Data webs sit in the centre of three service levels for data accessibility Ø Ingest-level services that receive and expose primary data Ø Ø n n Metadata aggregation services (data webs) Output-level services Further semantic processing can be undertaken by such output-level services, made possible by re-exposure of integrated metadata for programmatic access Within the biological realm, examples of such added-value output-level services that could be built on top of data webs for images could include: Ø Semantic slicing through the data, to present users with simpler topic-specific views Ø Use of Google Maps and other geospatial information for localization of wildlife photos Ø Automated programmatic use of the Rotterdam University gene name disambiguation service when dealing with gene expression images
Output-level services n Data webs sit in the centre of three service levels for data accessibility Ø Ingest-level services that receive and expose primary data Ø Ø n n Metadata aggregation services (data webs) Output-level services Further semantic processing can be undertaken by such output-level services, made possible by re-exposure of integrated metadata for programmatic access Within the biological realm, examples of such added-value output-level services that could be built on top of data webs for images could include: Ø Semantic slicing through the data, to present users with simpler topic-specific views Ø Use of Google Maps and other geospatial information for localization of wildlife photos Ø Automated programmatic use of the Rotterdam University gene name disambiguation service when dealing with gene expression images
 Acknowledgements Graham Klyne Chris Catton web For sponsoring this workshop N. B. We are likely to be appointing at least one new researcher within the Image Bioinformatics Research Group later this summer. Enquiries and CVs to david. shotton@zoo. ox. ac. uk.
Acknowledgements Graham Klyne Chris Catton web For sponsoring this workshop N. B. We are likely to be appointing at least one new researcher within the Image Bioinformatics Research Group later this summer. Enquiries and CVs to david. shotton@zoo. ox. ac. uk.
 Thank you for your attention Questions?
Thank you for your attention Questions?
 Other metadata aggregation projects n n The CNRI’s CORDRA Project (http: //www. cordra. net/) has specified an aggregation service similar in concept to our data web, whereby a search of federated metadata provides a means of searching multiple repositories The US Defence Department’s Defence Virtual Information Architecture (http: //www. cnri. reston. va. us/dtic. html) is based on the same CNRI system The a. DORe system (Van de Sompel et al. , 2005; http: //comjnl. oxfordjournals. org/cgi/rapidpdf/bxh 114 v 1. pdf) has practically demonstrated many of the CORDRA concepts These developments can inform and benefit our proposed data web development Ø However, they differ in that they involve active top-down federation of the participating repositories, whereas a data web simply requires repositories to expose their metadata on the Web Ø Furthermore, they do not employ Semantic Web technologies
Other metadata aggregation projects n n The CNRI’s CORDRA Project (http: //www. cordra. net/) has specified an aggregation service similar in concept to our data web, whereby a search of federated metadata provides a means of searching multiple repositories The US Defence Department’s Defence Virtual Information Architecture (http: //www. cnri. reston. va. us/dtic. html) is based on the same CNRI system The a. DORe system (Van de Sompel et al. , 2005; http: //comjnl. oxfordjournals. org/cgi/rapidpdf/bxh 114 v 1. pdf) has practically demonstrated many of the CORDRA concepts These developments can inform and benefit our proposed data web development Ø However, they differ in that they involve active top-down federation of the participating repositories, whereas a data web simply requires repositories to expose their metadata on the Web Ø Furthermore, they do not employ Semantic Web technologies


