0757fd95573839b5e8508d30e1187933.ppt
- Количество слайдов: 23
 Reporting. Tools: an automated result processing toolkit for high throughput genomic analyses Jessica Larson, Ph. D Computational Biologist Genentech, Inc. larson. jessica@gene. com July 19 2013 © 2013, Genentech
Reporting. Tools: an automated result processing toolkit for high throughput genomic analyses Jessica Larson, Ph. D Computational Biologist Genentech, Inc. larson. jessica@gene. com July 19 2013 © 2013, Genentech
 Outline (1) (2) (3) (4) (5) Reporting. Tools introduction and basics Reporting. Tools and microarray experiments Reporting. Tools and RNA-seq experiments Reporting. Tools and knitr Reporting. Tools and shiny 1
Outline (1) (2) (3) (4) (5) Reporting. Tools introduction and basics Reporting. Tools and microarray experiments Reporting. Tools and RNA-seq experiments Reporting. Tools and knitr Reporting. Tools and shiny 1
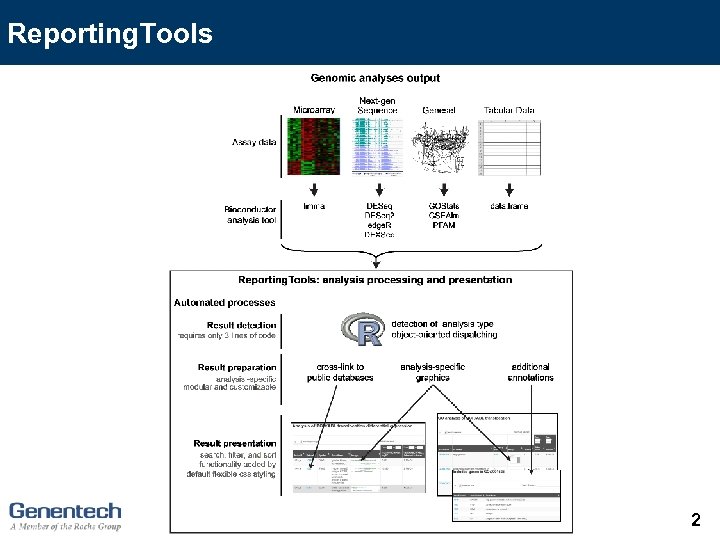 Reporting. Tools 2
Reporting. Tools 2
 Example output and code from the package vignettes http: //research-pub. gene. com/Reporting. Tools/ 3
Example output and code from the package vignettes http: //research-pub. gene. com/Reporting. Tools/ 3
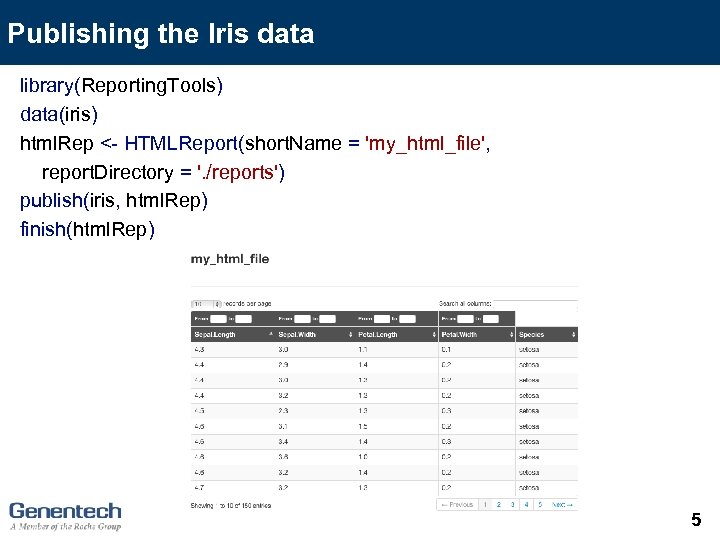
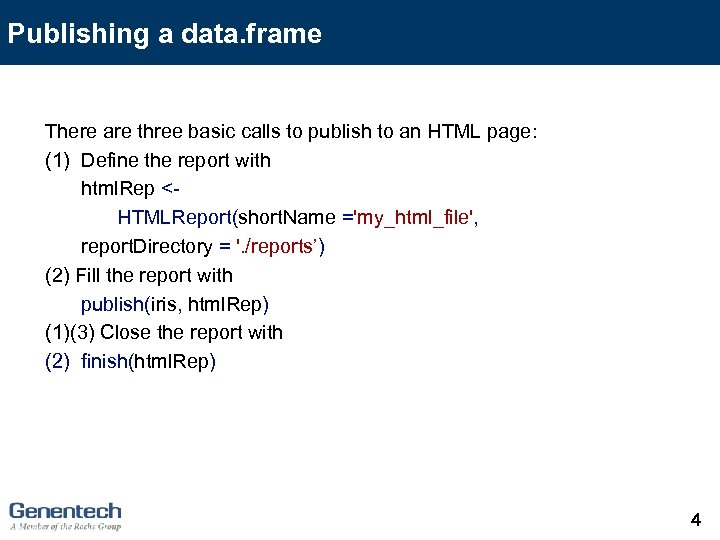 Publishing a data. frame There are three basic calls to publish to an HTML page: (1) Define the report with html. Rep
Publishing a data. frame There are three basic calls to publish to an HTML page: (1) Define the report with html. Rep
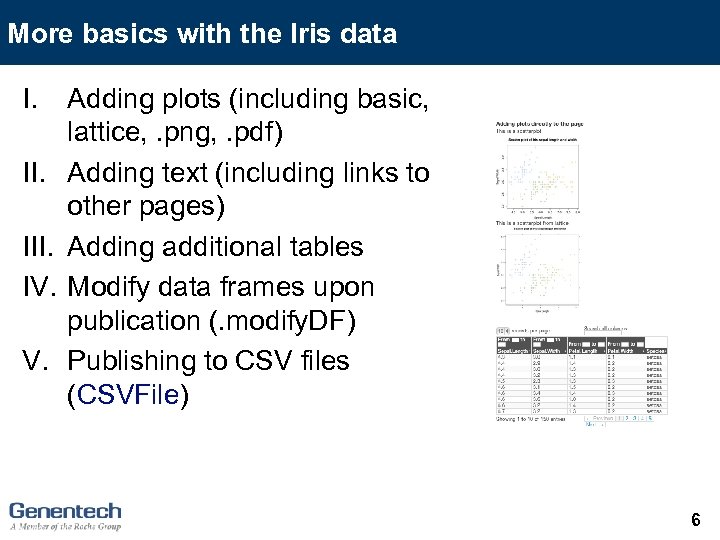 More basics with the Iris data I. III. IV. V. Adding plots (including basic, lattice, . png, . pdf) Adding text (including links to other pages) Adding additional tables Modify data frames upon publication (. modify. DF) Publishing to CSV files (CSVFile) 6
More basics with the Iris data I. III. IV. V. Adding plots (including basic, lattice, . png, . pdf) Adding text (including links to other pages) Adding additional tables Modify data frames upon publication (. modify. DF) Publishing to CSV files (CSVFile) 6
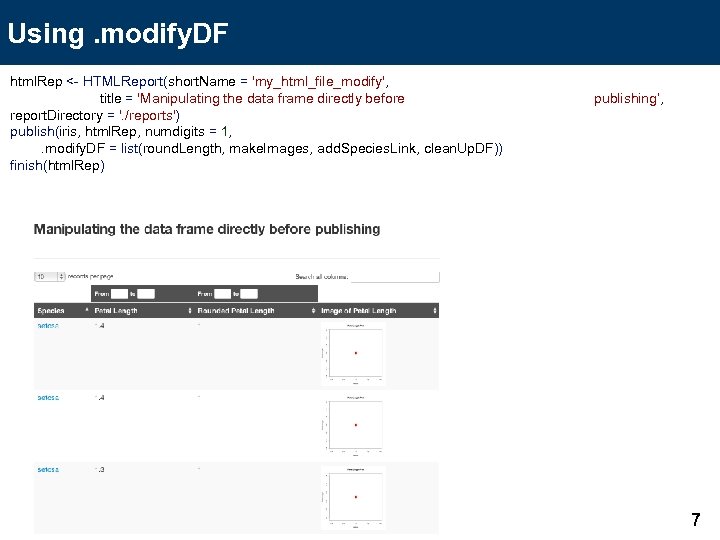 Using. modify. DF html. Rep <- HTMLReport(short. Name = 'my_html_file_modify', title = 'Manipulating the data frame directly before report. Directory = '. /reports') publish(iris, html. Rep, numdigits = 1, . modify. DF = list(round. Length, make. Images, add. Species. Link, clean. Up. DF)) finish(html. Rep) publishing’, 7
Using. modify. DF html. Rep <- HTMLReport(short. Name = 'my_html_file_modify', title = 'Manipulating the data frame directly before report. Directory = '. /reports') publish(iris, html. Rep, numdigits = 1, . modify. DF = list(round. Length, make. Images, add. Species. Link, clean. Up. DF)) finish(html. Rep) publishing’, 7
 Outline (1) (2) (3) (4) (5) Reporting. Tools introduction and basics Reporting. Tools and microarray experiments Reporting. Tools and RNA-seq experiments Reporting. Tools and knitr Reporting. Tools and shiny 8
Outline (1) (2) (3) (4) (5) Reporting. Tools introduction and basics Reporting. Tools and microarray experiments Reporting. Tools and RNA-seq experiments Reporting. Tools and knitr Reporting. Tools and shiny 8
 Microarray examples I. Publish output from limma (add new images and links) II. Publish GO and PFAM analysis output III. Publish gene sets IV. Create index pages 9
Microarray examples I. Publish output from limma (add new images and links) II. Publish GO and PFAM analysis output III. Publish gene sets IV. Create index pages 9
 Outline (1) (2) (3) (4) (5) Reporting. Tools introduction and basics Reporting. Tools and microarray experiments Reporting. Tools and RNA-seq experiments Reporting. Tools and knitr Reporting. Tools and shiny 10
Outline (1) (2) (3) (4) (5) Reporting. Tools introduction and basics Reporting. Tools and microarray experiments Reporting. Tools and RNA-seq experiments Reporting. Tools and knitr Reporting. Tools and shiny 10
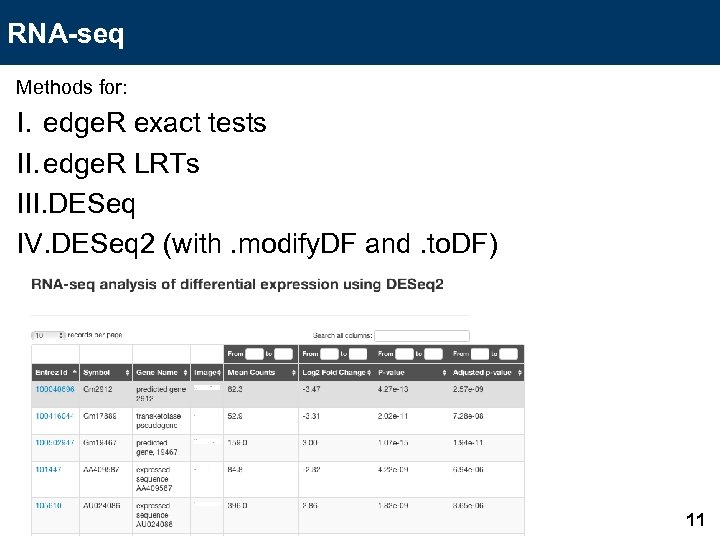 RNA-seq Methods for: I. edge. R exact tests II. edge. R LRTs III. DESeq IV. DESeq 2 (with. modify. DF and. to. DF) 11
RNA-seq Methods for: I. edge. R exact tests II. edge. R LRTs III. DESeq IV. DESeq 2 (with. modify. DF and. to. DF) 11
 Outline (1) (2) (3) (4) (5) Reporting. Tools introduction and basics Reporting. Tools and microarray experiments Reporting. Tools and RNA-seq experiments Reporting. Tools and knitr Reporting. Tools and shiny 12
Outline (1) (2) (3) (4) (5) Reporting. Tools introduction and basics Reporting. Tools and microarray experiments Reporting. Tools and RNA-seq experiments Reporting. Tools and knitr Reporting. Tools and shiny 12
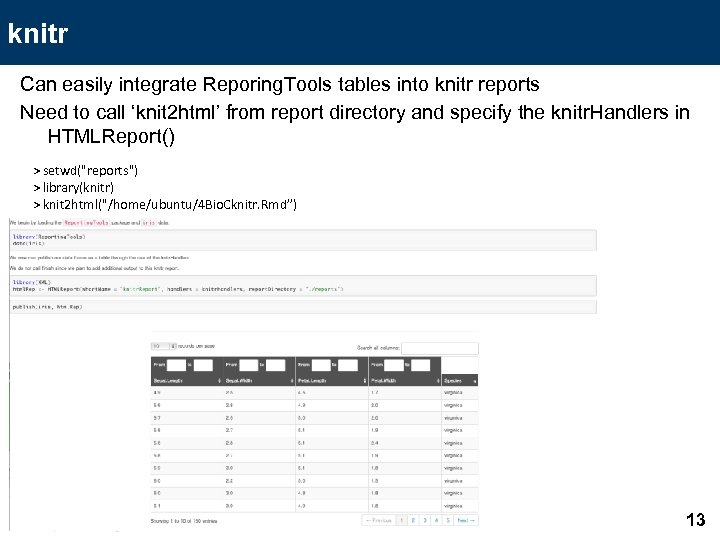 knitr Can easily integrate Reporing. Tools tables into knitr reports Need to call ‘knit 2 html’ from report directory and specify the knitr. Handlers in HTMLReport() > setwd("reports") > library(knitr) > knit 2 html("/home/ubuntu/4 Bio. Cknitr. Rmd”) 13
knitr Can easily integrate Reporing. Tools tables into knitr reports Need to call ‘knit 2 html’ from report directory and specify the knitr. Handlers in HTMLReport() > setwd("reports") > library(knitr) > knit 2 html("/home/ubuntu/4 Bio. Cknitr. Rmd”) 13
 Updates to run the knitr example (due to permission issues) (1) (2) (3) (4) Open 4 Bio. Cknitr. Rmd Save to the /home/ubuntu directory setwd(“reports”) Then call knit 2 html("/home/ubuntu/4 Bio. Cknitr. Rmd") 14
Updates to run the knitr example (due to permission issues) (1) (2) (3) (4) Open 4 Bio. Cknitr. Rmd Save to the /home/ubuntu directory setwd(“reports”) Then call knit 2 html("/home/ubuntu/4 Bio. Cknitr. Rmd") 14
 Outline (1) (2) (3) (4) (5) Reporting. Tools introduction and basics Reporting. Tools and microarray experiments Reporting. Tools and RNA-seq experiments Reporting. Tools and knitr Reporting. Tools and shiny 15
Outline (1) (2) (3) (4) (5) Reporting. Tools introduction and basics Reporting. Tools and microarray experiments Reporting. Tools and RNA-seq experiments Reporting. Tools and knitr Reporting. Tools and shiny 15
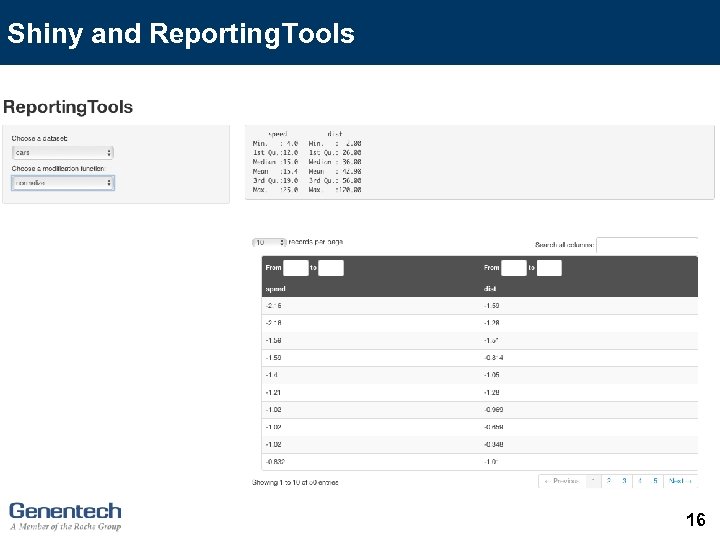 Shiny and Reporting. Tools 16
Shiny and Reporting. Tools 16
 Updates to run the shiny example (due to permission issues) (1) (2) (3) (4) Open server. R and ui. R Save these to the /home/ubuntu/reports directory setwd(“reports”) Then call my. Run. App() 17
Updates to run the shiny example (due to permission issues) (1) (2) (3) (4) Open server. R and ui. R Save these to the /home/ubuntu/reports directory setwd(“reports”) Then call my. Run. App() 17
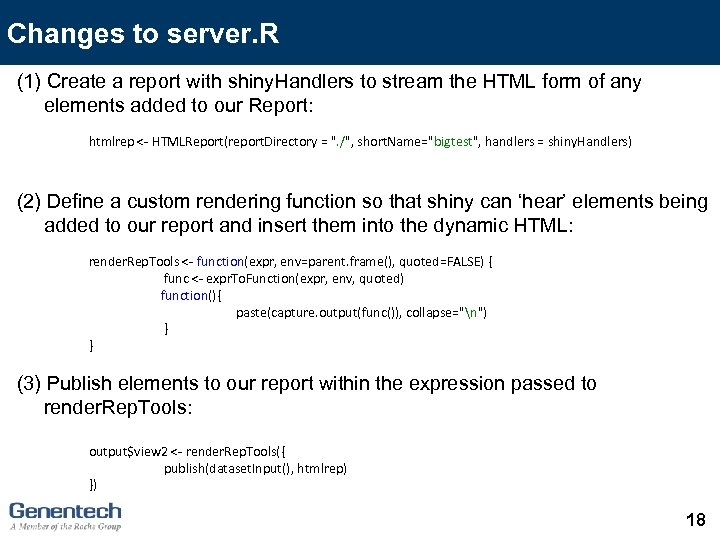 Changes to server. R (1) Create a report with shiny. Handlers to stream the HTML form of any elements added to our Report: htmlrep <- HTMLReport(report. Directory = ". /", short. Name="bigtest", handlers = shiny. Handlers) (2) Define a custom rendering function so that shiny can ‘hear’ elements being added to our report and insert them into the dynamic HTML: render. Rep. Tools <- function(expr, env=parent. frame(), quoted=FALSE) { func <- expr. To. Function(expr, env, quoted) function(){ paste(capture. output(func()), collapse="n") } } (3) Publish elements to our report within the expression passed to render. Rep. Tools: output$view 2 <- render. Rep. Tools({ publish(dataset. Input(), htmlrep) }) 18
Changes to server. R (1) Create a report with shiny. Handlers to stream the HTML form of any elements added to our Report: htmlrep <- HTMLReport(report. Directory = ". /", short. Name="bigtest", handlers = shiny. Handlers) (2) Define a custom rendering function so that shiny can ‘hear’ elements being added to our report and insert them into the dynamic HTML: render. Rep. Tools <- function(expr, env=parent. frame(), quoted=FALSE) { func <- expr. To. Function(expr, env, quoted) function(){ paste(capture. output(func()), collapse="n") } } (3) Publish elements to our report within the expression passed to render. Rep. Tools: output$view 2 <- render. Rep. Tools({ publish(dataset. Input(), htmlrep) }) 18
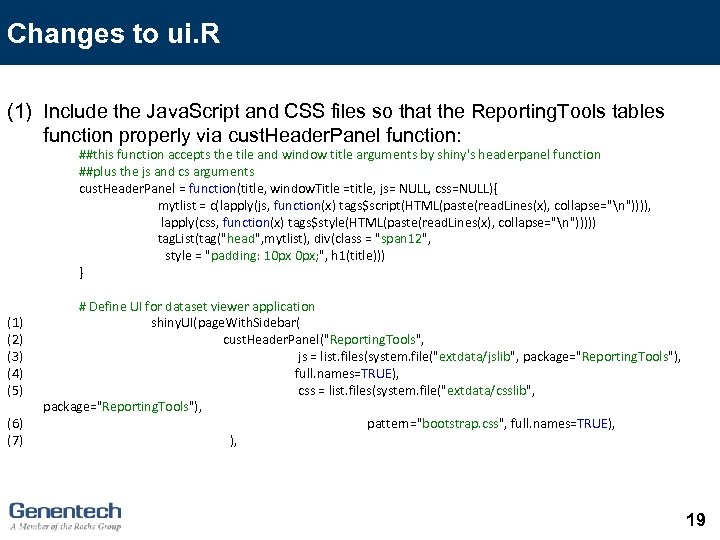 Changes to ui. R (1) Include the Java. Script and CSS files so that the Reporting. Tools tables function properly via cust. Header. Panel function: ##this function accepts the tile and window title arguments by shiny's headerpanel function ##plus the js and cs arguments cust. Header. Panel = function(title, window. Title =title, js= NULL, css=NULL){ mytlist = c(lapply(js, function(x) tags$script(HTML(paste(read. Lines(x), collapse="n")))), lapply(css, function(x) tags$style(HTML(paste(read. Lines(x), collapse="n"))))) tag. List(tag("head", mytlist), div(class = "span 12", style = "padding: 10 px 0 px; ", h 1(title))) } (1) (2) (3) (4) (5) (6) (7) # Define UI for dataset viewer application shiny. UI(page. With. Sidebar( cust. Header. Panel("Reporting. Tools", js = list. files(system. file("extdata/jslib", package="Reporting. Tools"), full. names=TRUE), css = list. files(system. file("extdata/csslib", package="Reporting. Tools"), pattern="bootstrap. css", full. names=TRUE), ), 19
Changes to ui. R (1) Include the Java. Script and CSS files so that the Reporting. Tools tables function properly via cust. Header. Panel function: ##this function accepts the tile and window title arguments by shiny's headerpanel function ##plus the js and cs arguments cust. Header. Panel = function(title, window. Title =title, js= NULL, css=NULL){ mytlist = c(lapply(js, function(x) tags$script(HTML(paste(read. Lines(x), collapse="n")))), lapply(css, function(x) tags$style(HTML(paste(read. Lines(x), collapse="n"))))) tag. List(tag("head", mytlist), div(class = "span 12", style = "padding: 10 px 0 px; ", h 1(title))) } (1) (2) (3) (4) (5) (6) (7) # Define UI for dataset viewer application shiny. UI(page. With. Sidebar( cust. Header. Panel("Reporting. Tools", js = list. files(system. file("extdata/jslib", package="Reporting. Tools"), full. names=TRUE), css = list. files(system. file("extdata/csslib", package="Reporting. Tools"), pattern="bootstrap. css", full. names=TRUE), ), 19
 Changes to ui. R (2) Declare elements formated by Reporting. Tools as html. Output main. Panel( ) verbatim. Text. Output("summary"), html. Output("view 2") This indicates to the shiny system that the output with be HTML code 20
Changes to ui. R (2) Declare elements formated by Reporting. Tools as html. Output main. Panel( ) verbatim. Text. Output("summary"), html. Output("view 2") This indicates to the shiny system that the output with be HTML code 20
 Future methods DESeq 2 methods Return the decorated DF More flexibility with annotations 21
Future methods DESeq 2 methods Return the decorated DF More flexibility with annotations 21
 Acknowledgements Jason Hackney Josh Kaminker Melanie Huntley Christina Chaivorapol Gabriel Becker Michael Lawrence Robert Gentleman Martin Morgan Dan Tenenbaum 22
Acknowledgements Jason Hackney Josh Kaminker Melanie Huntley Christina Chaivorapol Gabriel Becker Michael Lawrence Robert Gentleman Martin Morgan Dan Tenenbaum 22