3f91bc97cc40f030cfaba2f2f66b40fa.ppt
- Количество слайдов: 14

Reforming Reactome Robin Haw 15 -19 th August 2012 COMBINE – Toronto Meeting www. reactome. org
What is Reactome? • Open source and open access pathway database – 1300+ human pathways encompassing metabolism, signaling, gene regulation, and other biological processes • Extensively cross-referenced to external bioinformatics databases • Tools and datasets for browsing and visualizing pathway data, and interpreting your experimental data. www. reactome. org
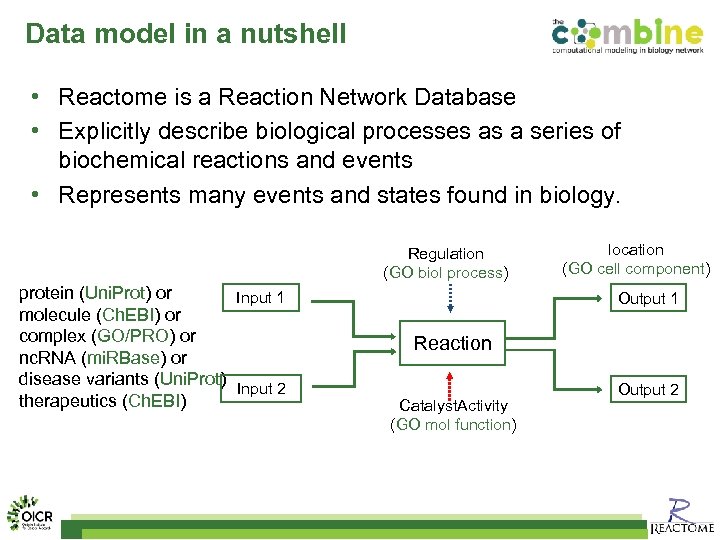
Data model in a nutshell • Reactome is a Reaction Network Database • Explicitly describe biological processes as a series of biochemical reactions and events • Represents many events and states found in biology. Regulation (GO biol process) protein (Uni. Prot) or Input 1 molecule (Ch. EBI) or complex (GO/PRO) or nc. RNA (mi. RBase) or disease variants (Uni. Prot) Input 2 therapeutics (Ch. EBI) location (GO cell component) Output 1 Reaction Catalyst. Activity (GO mol function) Output 2
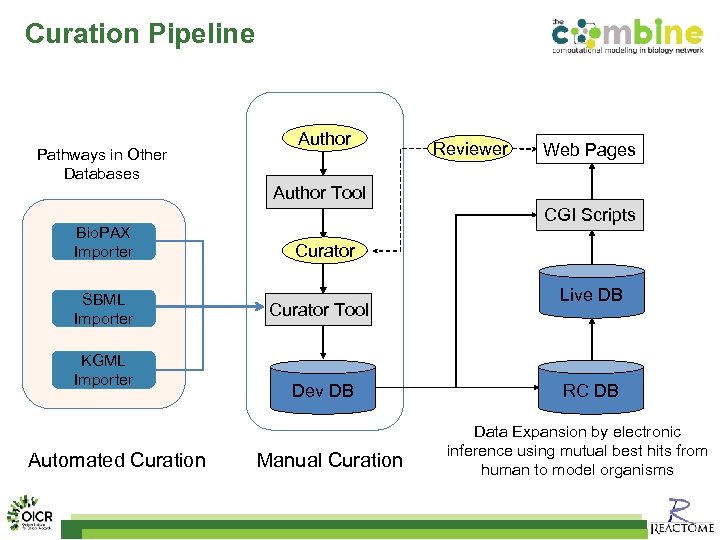
Curation Pipeline Pathways in Other Databases Bio. PAX Importer SBML Importer KGML Importer Automated Curation Author Reviewer Web Pages Author Tool CGI Scripts Curator Tool Dev DB Manual Curation Live DB RC DB Data Expansion by electronic inference using mutual best hits from human to model organisms
Focus on Disease Topics • Not new to Reactome! • Reorganized the Pathway Hierarchy. • Modified the Data Model. • Updating the Pathway Browser. • Annotate: • An infection introduces new proteins into the cell. • The amount of a normal protein is changed and this changes the function of the protein. • A mutation (somatic or germline) changes the function of a protein. • Mode of action of anti-cancer therapeutics and antibodies.
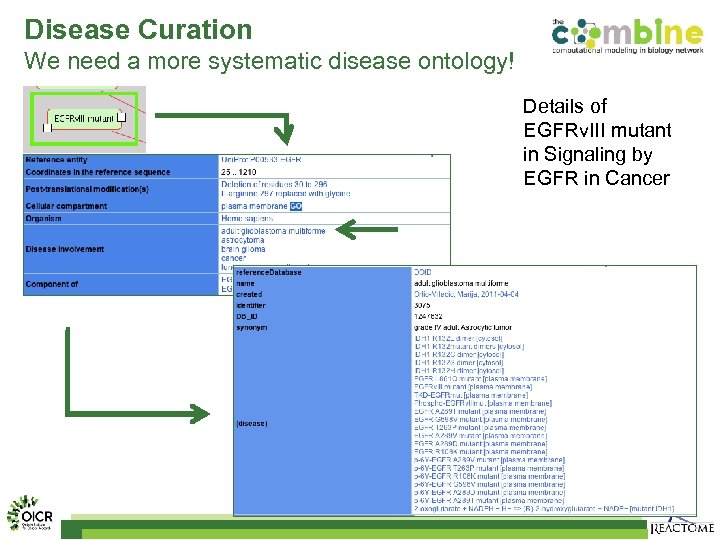
Disease Curation We need a more systematic disease ontology! Details of EGFRv. III mutant in Signaling by EGFR in Cancer
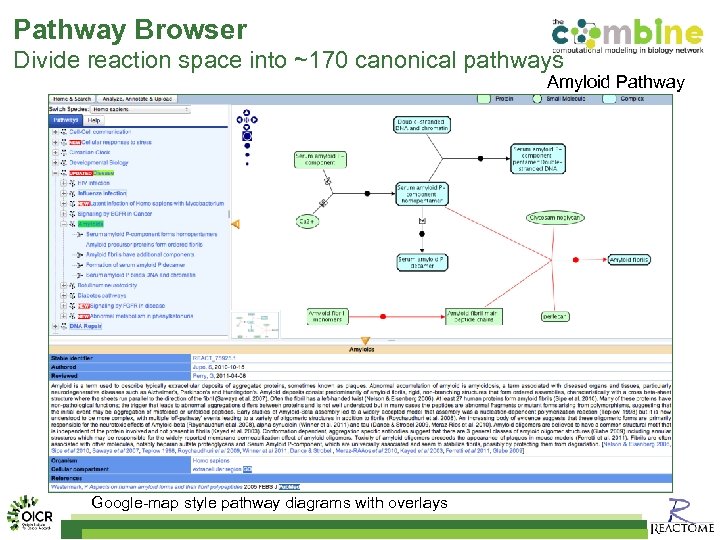
Pathway Browser Divide reaction space into ~170 canonical pathways Amyloid Pathway Google-map style pathway diagrams with overlays
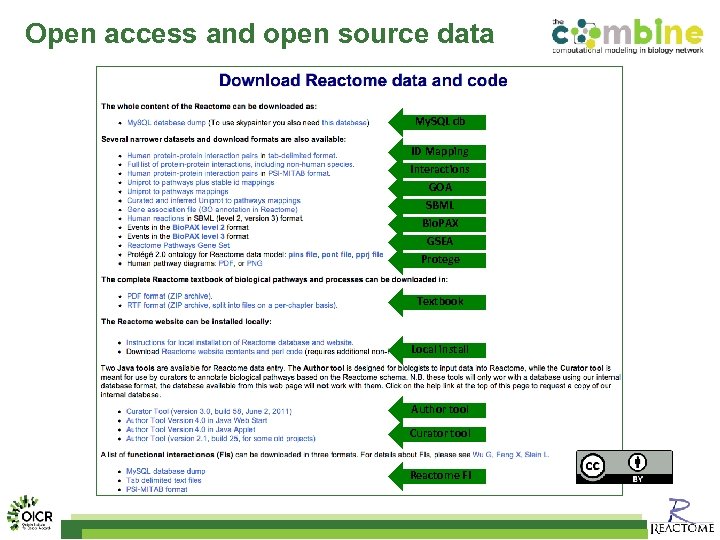
Open access and open source data My. SQL db ID Mapping Interactions GOA SBML Bio. PAX GSEA Protege Textbook Local install Author tool Curator tool Reactome FI
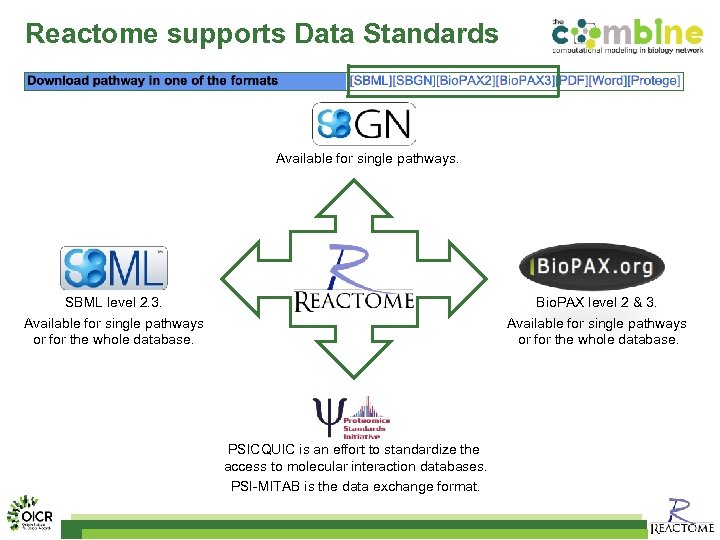
Reactome supports Data Standards Available for single pathways. Bio. PAX level 2 & 3. Available for single pathways or for the whole database. SBML level 2. 3. Available for single pathways or for the whole database. PSICQUIC is an effort to standardize the access to molecular interaction databases. PSI-MITAB is the data exchange format.
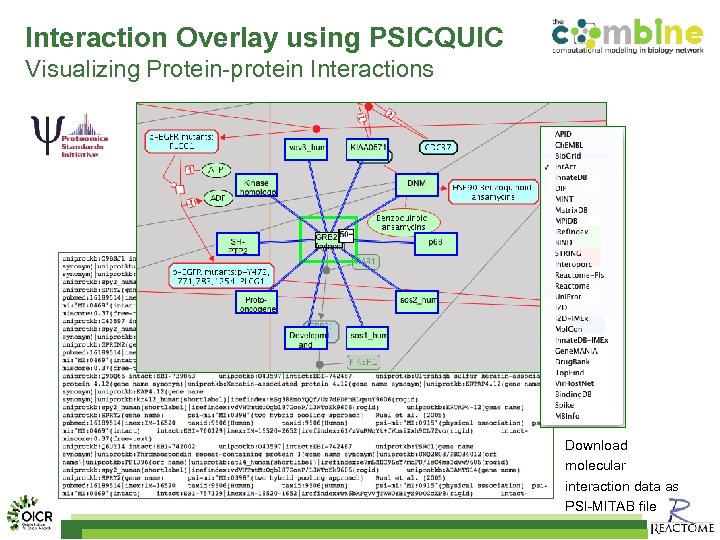
Interaction Overlay using PSICQUIC Visualizing Protein-protein Interactions Download molecular interaction data as PSI-MITAB file
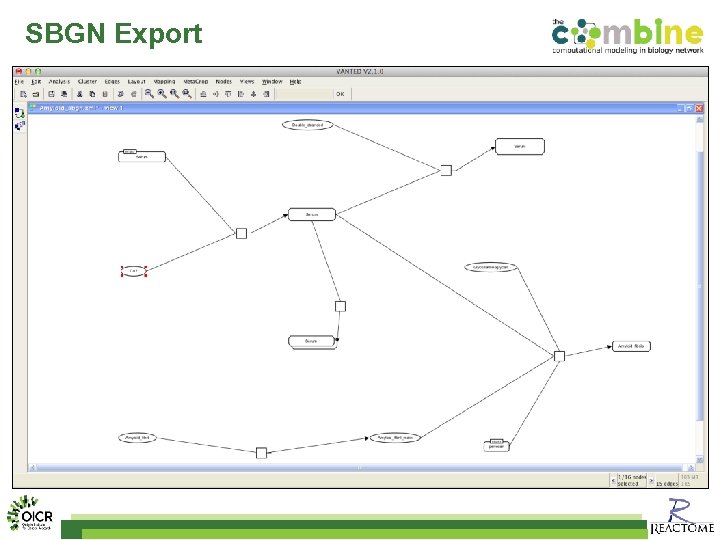
SBGN Export
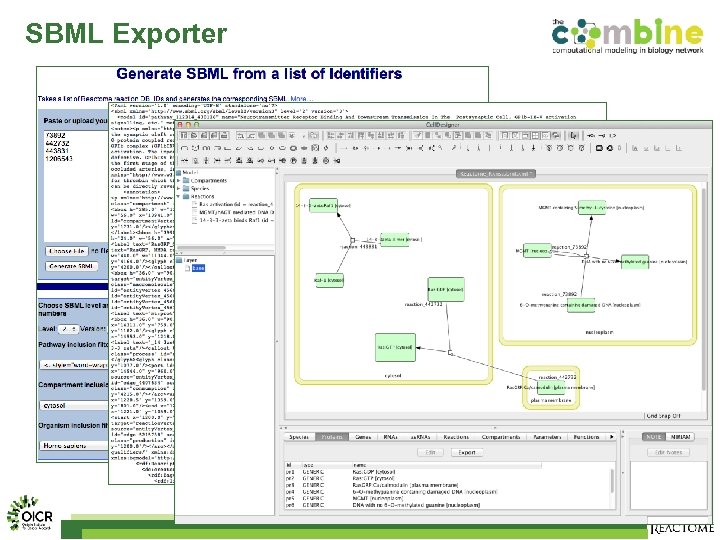
SBML Exporter

Continuing Priorities • Increase the number of curated proteins and other functional entities. • Supplement normal pathways with variant reactions representing disease states. • Improve annotation consistency and enrich the data model. • Continued support for SBGN, SBML, Bio. PAX and PSI. • Enhance the web site and other resources to meet the needs of a growing and diverse user community.

Acknowledgements • • • Marija Orlic-Milacic Karen Rothfels Lisa Matthews Marc Gillespie Guanming Wu Irina Kalatskaya Christina Yung Michael Caudy David Croft Adrian Duong Phani Garapati Bijay Jassal • • • Steve Jupe Bruce May Antonio Fabregat Mundo Veronica Shamovsky Heeyeon Song Joel Weiser Mark Williams Henning Hermjakob Peter D’Eustachio Lincoln Stein Ministry of Economic Development and Innovation
3f91bc97cc40f030cfaba2f2f66b40fa.ppt