e188fc7eec78ff90229ef84d21047856.ppt
- Количество слайдов: 34

Recombination Mapping

Why • • A fundamental problem in human genetics today is locating and identifying the specific gene responsible for a given genetic disease. However, the disease is just a phenotype, and gene responsible for that phenotype might be very different from what we would expect. – For instance, Lesch-Nyhan syndrome’s most spectacular manifestation is self-mutilating behavior. The Lesch-Nyhan gene codes for hypoxanthine-guanine phosphoribosyl transferase, which helps salvage nucleotides derived from the breakdown of nucleic acids. • So, we need to reduce the number of candidate genes to a manageable level. • Using the naturally occurring recombination process to map genes remains the best way to localize the gene responsible for a genetic disease. The goal is to reduce the amount of DNA that need to be searched to a small region, a few million base pairs or so. Below that level, molecular tools need to be employed.

Markers for Mapping • What makes a good marker: – co-dominant (so homozygotes and heterozygotes can be distinguished) – many alleles at each locus (so most people will be heterozygous and different from each other) – many loci well distributed throughout the genome – easy to detect, especially with automated machinery • No system is perfect
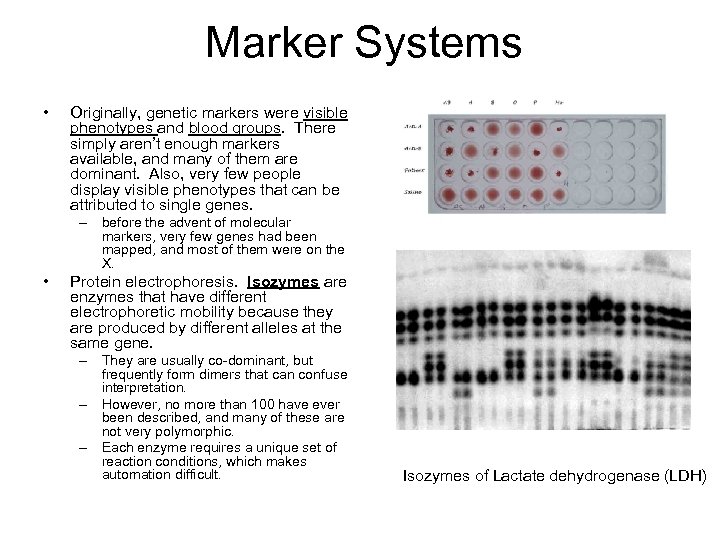
Marker Systems • Originally, genetic markers were visible phenotypes and blood groups. There simply aren’t enough markers available, and many of them are dominant. Also, very few people display visible phenotypes that can be attributed to single genes. – before the advent of molecular markers, very few genes had been mapped, and most of them were on the X. • Protein electrophoresis. Isozymes are enzymes that have different electrophoretic mobility because they are produced by different alleles at the same gene. – They are usually co-dominant, but frequently form dimers that can confuse interpretation. – However, no more than 100 have ever been described, and many of these are not very polymorphic. – Each enzyme requires a unique set of reaction conditions, which makes automation difficult. Isozymes of Lactate dehydrogenase (LDH)
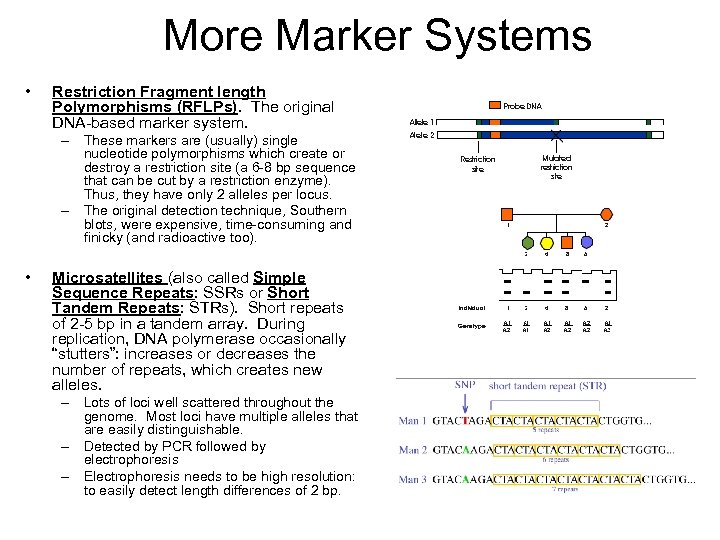
More Marker Systems • Restriction Fragment length Polymorphisms (RFLPs). The original DNA-based marker system. – These markers are (usually) single nucleotide polymorphisms which create or destroy a restriction site (a 6 -8 bp sequence that can be cut by a restriction enzyme). Thus, they have only 2 alleles per locus. – The original detection technique, Southern blots, were expensive, time-consuming and finicky (and radioactive too). • Microsatellites (also called Simple Sequence Repeats: SSRs or Short Tandem Repeats: STRs). Short repeats of 2 -5 bp in a tandem array. During replication, DNA polymerase occasionally “stutters”: increases or decreases the number of repeats, which creates new alleles. – Lots of loci well scattered throughout the genome. Most loci have multiple alleles that are easily distinguishable. – Detected by PCR followed by electrophoresis – Electrophoresis needs to be high resolution: to easily detect length differences of 2 bp.
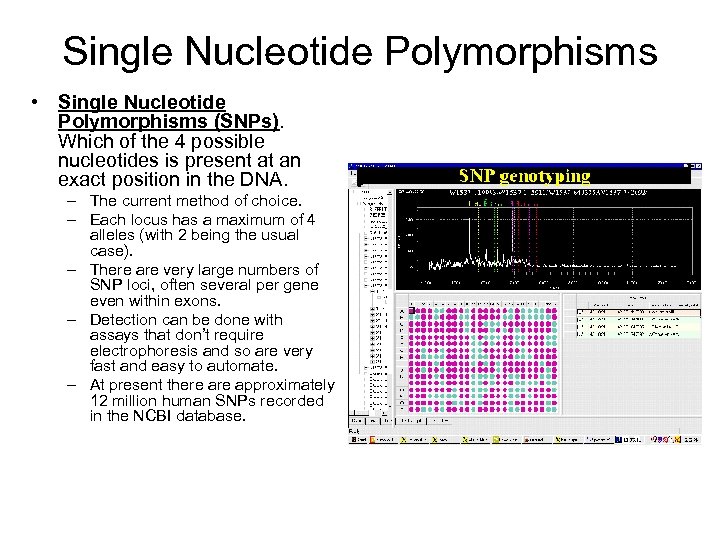
Single Nucleotide Polymorphisms • Single Nucleotide Polymorphisms (SNPs). Which of the 4 possible nucleotides is present at an exact position in the DNA. – The current method of choice. – Each locus has a maximum of 4 alleles (with 2 being the usual case). – There are very large numbers of SNP loci, often several per gene even within exons. – Detection can be done with assays that don’t require electrophoresis and so are very fast and easy to automate. – At present there approximately 12 million human SNPs recorded in the NCBI database.
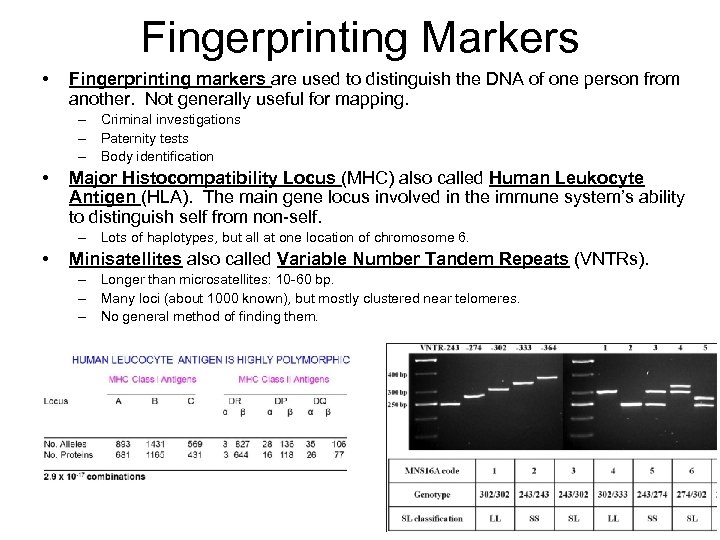
Fingerprinting Markers • Fingerprinting markers are used to distinguish the DNA of one person from another. Not generally useful for mapping. – Criminal investigations – Paternity tests – Body identification • Major Histocompatibility Locus (MHC) also called Human Leukocyte Antigen (HLA). The main gene locus involved in the immune system’s ability to distinguish self from non-self. – Lots of haplotypes, but all at one location of chromosome 6. • Minisatellites also called Variable Number Tandem Repeats (VNTRs). – Longer than microsatellites: 10 -60 bp. – Many loci (about 1000 known), but mostly clustered near telomeres. – No general method of finding them.
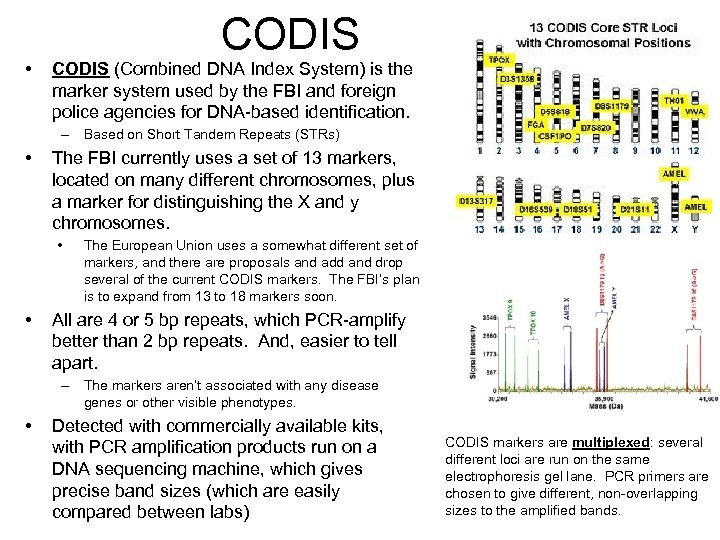
• CODIS (Combined DNA Index System) is the marker system used by the FBI and foreign police agencies for DNA-based identification. – Based on Short Tandem Repeats (STRs) • The FBI currently uses a set of 13 markers, located on many different chromosomes, plus a marker for distinguishing the X and y chromosomes. • • The European Union uses a somewhat different set of markers, and there are proposals and add and drop several of the current CODIS markers. The FBI’s plan is to expand from 13 to 18 markers soon. All are 4 or 5 bp repeats, which PCR-amplify better than 2 bp repeats. And, easier to tell apart. – The markers aren’t associated with any disease genes or other visible phenotypes. • Detected with commercially available kits, with PCR amplification products run on a DNA sequencing machine, which gives precise band sizes (which are easily compared between labs) CODIS markers are multiplexed: several different loci are run on the same electrophoresis gel lane. PCR primers are chosen to give different, non-overlapping sizes to the amplified bands.
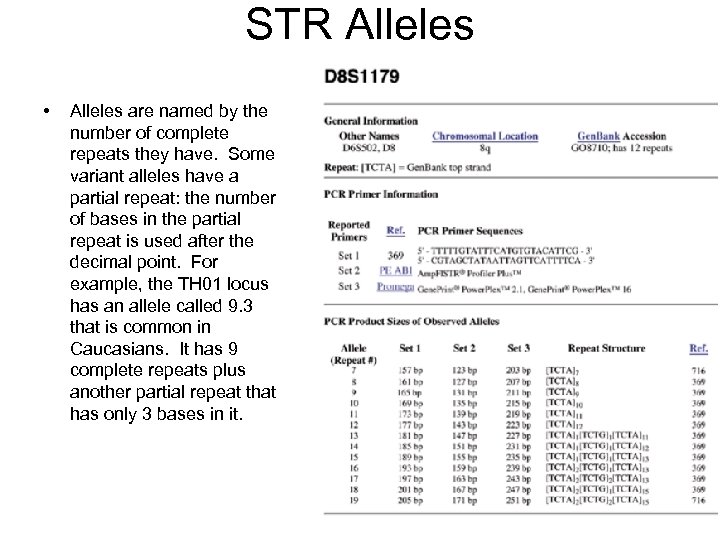
STR Alleles • Alleles are named by the number of complete repeats they have. Some variant alleles have a partial repeat: the number of bases in the partial repeat is used after the decimal point. For example, the TH 01 locus has an allele called 9. 3 that is common in Caucasians. It has 9 complete repeats plus another partial repeat that has only 3 bases in it.
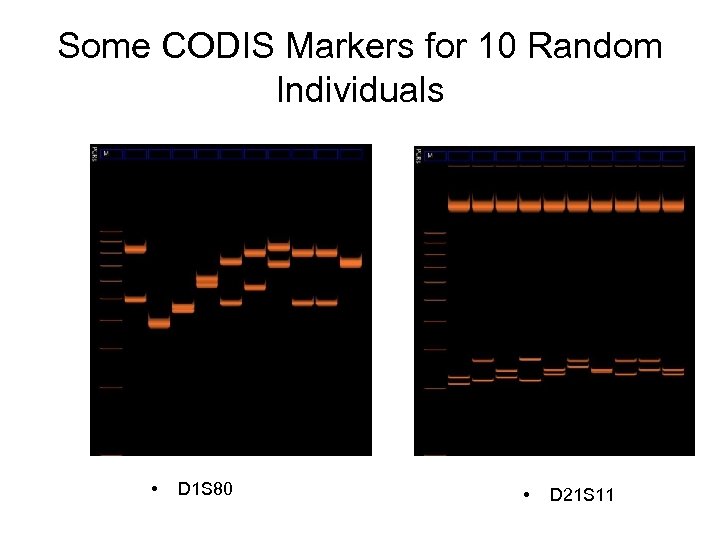
Some CODIS Markers for 10 Random Individuals • D 1 S 80 • D 21 S 11

Probability of Identity • The fundamental question with fingerprinting: what is the chance that two unrelated individuals will have the same genotype? (Probability of identity, Pi) – More alleles at any given locus improves the chances of not having unrelated people matching. – Since loci are genetically independent, Pi for several loci together is just the product of the individual Pi’s. – For perspective: there about 7 x 109 people living today, which means there about 25 x 1018 possible pairs of individuals. To be sure that you don’t misidentify someone, you need a Pi that is much less than 2. 5 x 10 -19. • Study done by National Institute of Standards and Technology (NIST) in 2012. – Examined 1036 unrelated individuals from the US, divided into these groups: Caucasian, African-American, Hispanic, and Asian. Ethnicity was self-identified, a procedure that obviously has some issues.
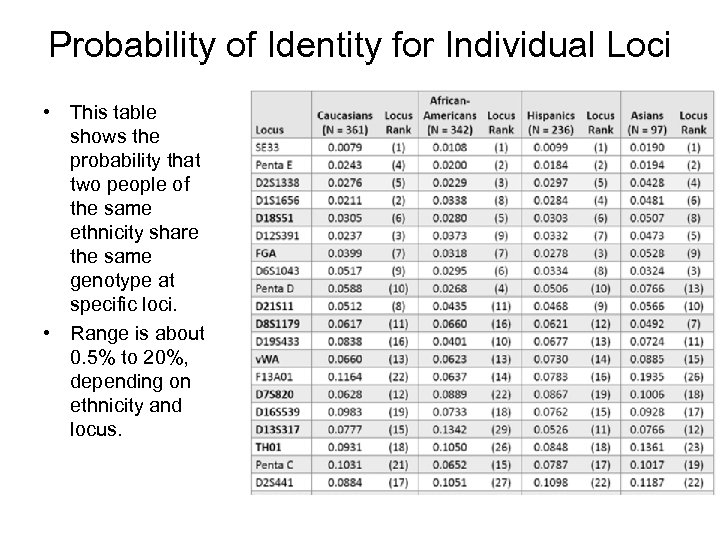
Probability of Identity for Individual Loci • This table shows the probability that two people of the same ethnicity share the same genotype at specific loci. • Range is about 0. 5% to 20%, depending on ethnicity and locus.
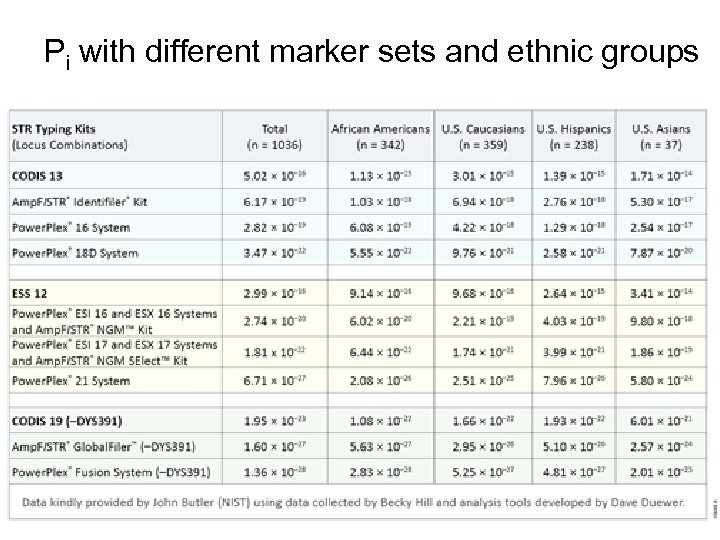
Pi with different marker sets and ethnic groups
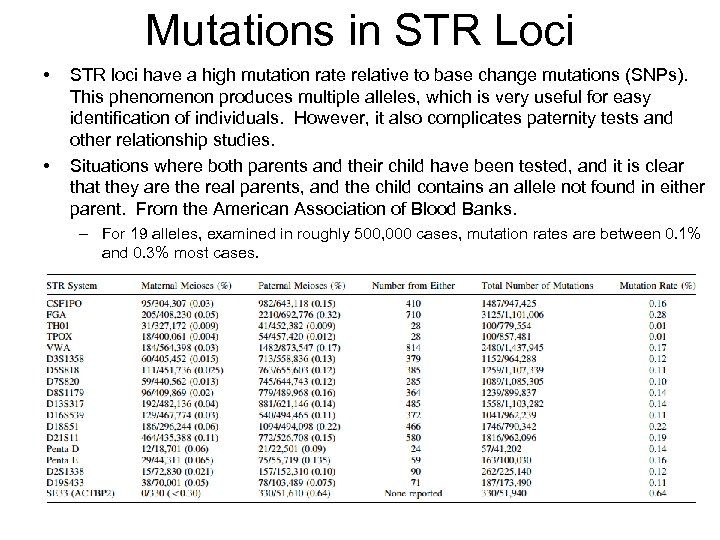
Mutations in STR Loci • • STR loci have a high mutation rate relative to base change mutations (SNPs). This phenomenon produces multiple alleles, which is very useful for easy identification of individuals. However, it also complicates paternity tests and other relationship studies. Situations where both parents and their child have been tested, and it is clear that they are the real parents, and the child contains an allele not found in either parent. From the American Association of Blood Banks. – For 19 alleles, examined in roughly 500, 000 cases, mutation rates are between 0. 1% and 0. 3% most cases.
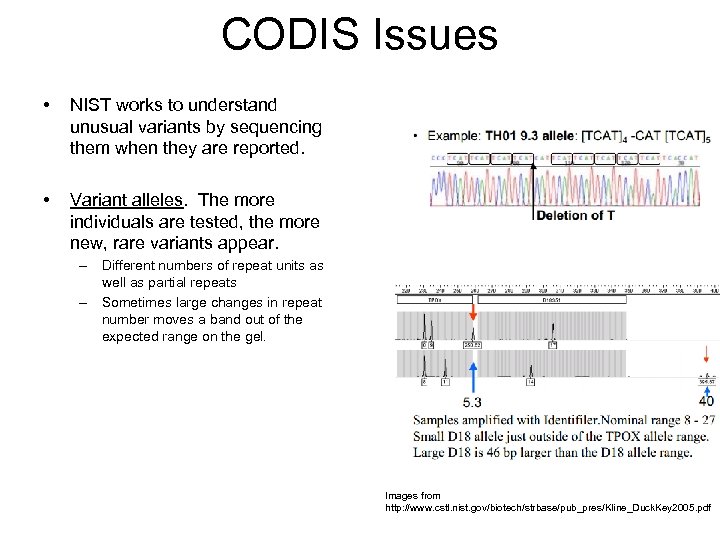
CODIS Issues • NIST works to understand unusual variants by sequencing them when they are reported. • Variant alleles. The more individuals are tested, the more new, rare variants appear. – Different numbers of repeat units as well as partial repeats – Sometimes large changes in repeat number moves a band out of the expected range on the gel. Images from http: //www. cstl. nist. gov/biotech/strbase/pub_pres/Kline_Duck. Key 2005. pdf
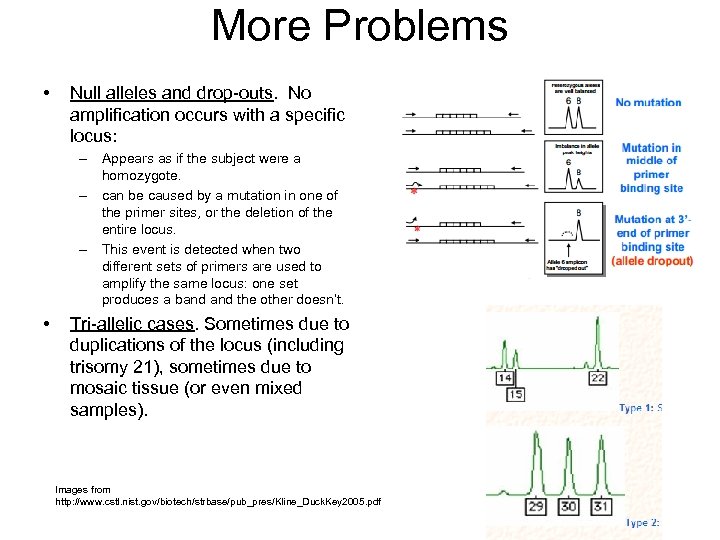
More Problems • Null alleles and drop-outs. No amplification occurs with a specific locus: – Appears as if the subject were a homozygote. – can be caused by a mutation in one of the primer sites, or the deletion of the entire locus. – This event is detected when two different sets of primers are used to amplify the same locus: one set produces a band the other doesn’t. • Tri-allelic cases. Sometimes due to duplications of the locus (including trisomy 21), sometimes due to mosaic tissue (or even mixed samples). Images from http: //www. cstl. nist. gov/biotech/strbase/pub_pres/Kline_Duck. Key 2005. pdf

Ethnicity Prediction • • Some loci have very different frequencies in different ethnic groups However, self-reported ethnicity isn’t very reliable. And, ethnicity isn’t a well-defined concept anyway. Mutation rates in STRs: identity by state (2 people have the same allele) vs. identity by descent (2 people have inherited an allele from the same common ancestor). • SNPs and Alu element insertions are more stable than STRs and probably work better for ethnicity prediction. • A related issue: linkage to disease genes. A DNA profile may give information about susceptibility to diseases. – Note that many disease genes were mapped using STR markers

Gene Mapping
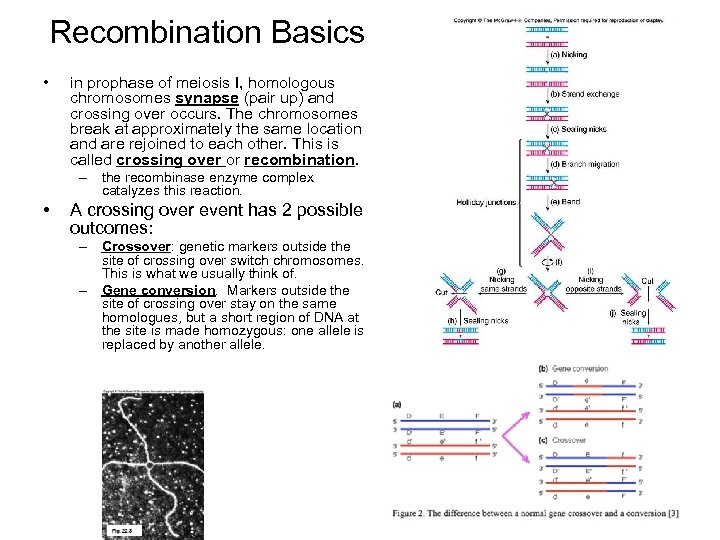
Recombination Basics • in prophase of meiosis I, homologous chromosomes synapse (pair up) and crossing over occurs. The chromosomes break at approximately the same location and are rejoined to each other. This is called crossing over or recombination. – the recombinase enzyme complex catalyzes this reaction. • A crossing over event has 2 possible outcomes: – Crossover: genetic markers outside the site of crossing over switch chromosomes. This is what we usually think of. – Gene conversion. Markers outside the site of crossing over stay on the same homologues, but a short region of DNA at the site is made homozygous: one allele is replaced by another allele.
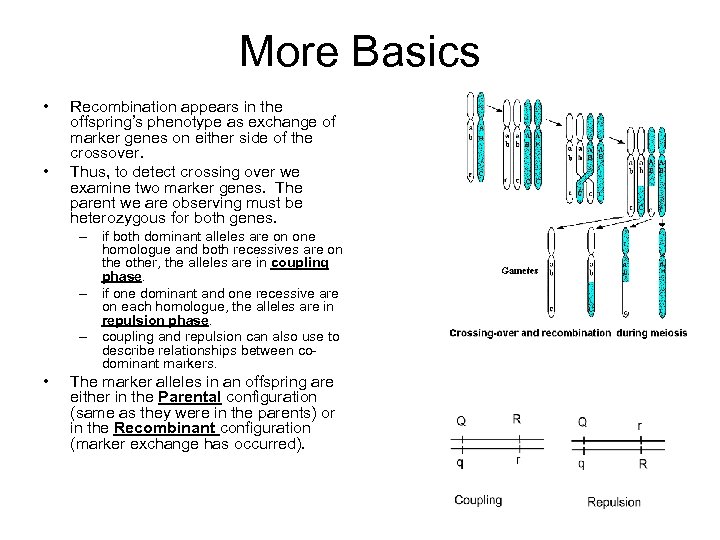
More Basics • • Recombination appears in the offspring’s phenotype as exchange of marker genes on either side of the crossover. Thus, to detect crossing over we examine two marker genes. The parent we are observing must be heterozygous for both genes. – if both dominant alleles are on one homologue and both recessives are on the other, the alleles are in coupling phase. – if one dominant and one recessive are on each homologue, the alleles are in repulsion phase. – coupling and repulsion can also use to describe relationships between codominant markers. • The marker alleles in an offspring are either in the Parental configuration (same as they were in the parents) or in the Recombinant configuration (marker exchange has occurred).
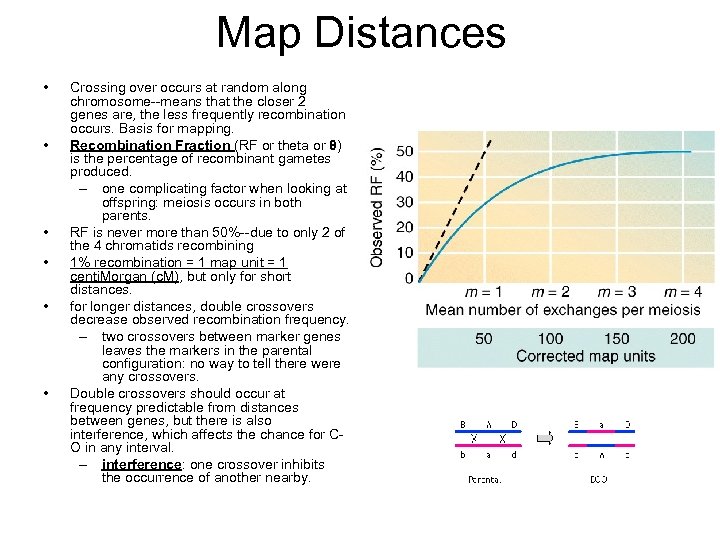
Map Distances • • • Crossing over occurs at random along chromosome--means that the closer 2 genes are, the less frequently recombination occurs. Basis for mapping. Recombination Fraction (RF or theta or θ) is the percentage of recombinant gametes produced. – one complicating factor when looking at offspring: meiosis occurs in both parents. RF is never more than 50%--due to only 2 of the 4 chromatids recombining 1% recombination = 1 map unit = 1 centi. Morgan (c. M), but only for short distances. for longer distances, double crossovers decrease observed recombination frequency. – two crossovers between marker genes leaves the markers in the parental configuration: no way to tell there were any crossovers. Double crossovers should occur at frequency predictable from distances between genes, but there is also interference, which affects the chance for CO in any interval. – interference: one crossover inhibits the occurrence of another nearby.

Mapping Function • • We want a gene map to be calibrated in map units that accurately reflect the frequency of crossovers between genes. The equation used to convert the observed recombination fraction into map units is called the mapping function. For a simple model of randomly placed crossovers and no interference, Haldane’s function works well: – • w = - ½ ln(1 -2θ) , where w is map distance and θ is the observed proportion of recombinants this expression produces the curve on the previous slide Interference complicates things, and a variety of functions can be used. Kosambi’s function is a common one: w = ¼ ln[(1+2θ) / (1 -2θ)] • • • Interference has been estimated for human genes, and it seems to be a very small effect. For a 10 c. M interval, only 0. 01% of the potential crossovers is inhibited by interference. Also, from a practical point of view, the main value of recombination mapping is finding a small region of DNA to search with molecular tools. Worrying about interference seems (to me) to be a lot of work for very little benefit. Further, it is clear that a crossover is not equally probable at every nucleotide: at the level of the DNA sequence, recombination primarily occurs at hot spots with very little in between:

Chiasmata • • • Crossing over is visible in the microscope as chiasmata (which is the plural form of chiasma). It is possible to count chiasmata. Each one counts as 50 map units (one crossover between 2 of the 4 DNA molecules at prophase of meiosis 1). In male meiosis (testicular biopsy), one study showed an average of 50. 6 chiasmata per cell. Multiplying by 50, this gives 2530 c. M as the length of the genetic map in males. In female meiosis (between 16 and 24 weeks of fetal life), an average of 70. 3 chiasmata per cell were seen. This gives a female map of 3515 c. M. Recombination mapping has given estimates of 2590 c. M for males and 4281 for females. So, females have more crossovers and a larger map than males. The total map length in humans is about 3000 c. M.

LOD Score Mapping • The general problems with mapping genes in humans: small families, uncontrolled matings, uncertain paternity. – Thus you can’t set up a test cross, where one parent is a heterozygote and the other is homozygous for other alleles, and count parental and recombinant offspring. • • • Given a pedigree family, the LOD score method involves determining the probability (the likelihood) of that family at different values of θ, the recombinant fraction. Then, the method allows you to add probabilities across different families, even if some information about them is missing or ambiguous. Also, each family can start with different parental arrangements of markers, and can have different numbers and types of children. The LOD score method is an example of a maximum likelihood procedure. The point of the maximum likelihood procedure is to estimate the value of a parameter that can’t be directly observed, in this case the recombination fraction. The likelihood (probability) of an observed set of data (the phenotypes seen in a family, in this case) is calculated as a function of that parameter. The parameter value that gives the maximum likelihood is taken as the best estimate of the parameter.
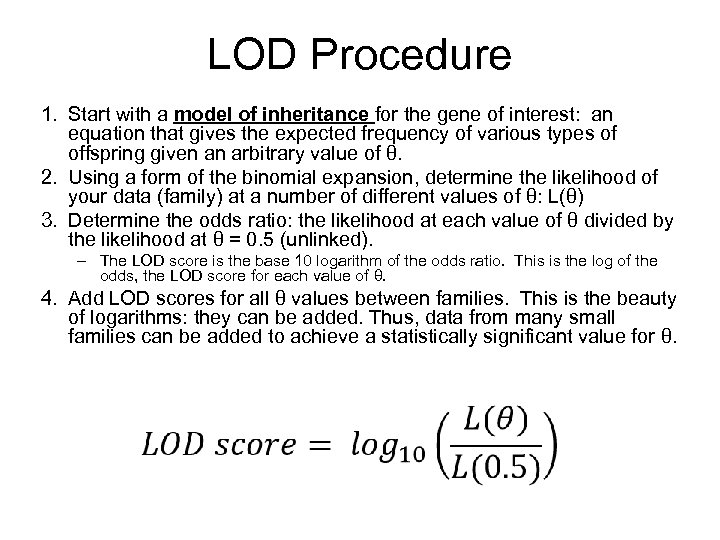
LOD Procedure 1. Start with a model of inheritance for the gene of interest: an equation that gives the expected frequency of various types of offspring given an arbitrary value of θ. 2. Using a form of the binomial expansion, determine the likelihood of your data (family) at a number of different values of θ: L(θ) 3. Determine the odds ratio: the likelihood at each value of θ divided by the likelihood at θ = 0. 5 (unlinked). – The LOD score is the base 10 logarithm of the odds ratio. This is the log of the odds, the LOD score for each value of θ. 4. Add LOD scores for all θ values between families. This is the beauty of logarithms: they can be added. Thus, data from many small families can be added to achieve a statistically significant value for θ.
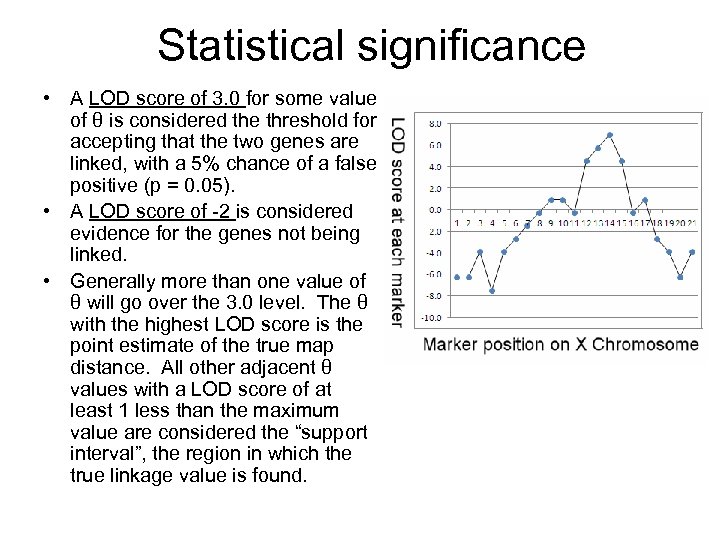
Statistical significance • A LOD score of 3. 0 for some value of θ is considered the threshold for accepting that the two genes are linked, with a 5% chance of a false positive (p = 0. 05). • A LOD score of -2 is considered evidence for the genes not being linked. • Generally more than one value of θ will go over the 3. 0 level. The θ with the highest LOD score is the point estimate of the true map distance. All other adjacent θ values with a LOD score of at least 1 less than the maximum value are considered the “support interval”, the region in which the true linkage value is found.
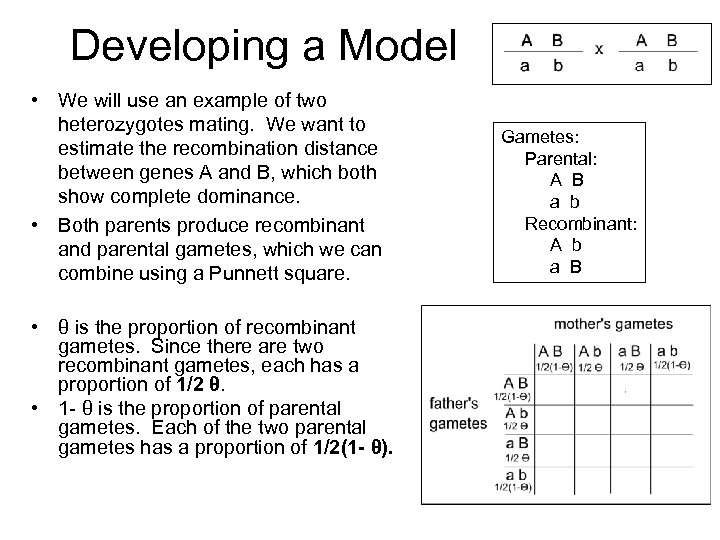
Developing a Model • We will use an example of two heterozygotes mating. We want to estimate the recombination distance between genes A and B, which both show complete dominance. • Both parents produce recombinant and parental gametes, which we can combine using a Punnett square. • θ is the proportion of recombinant gametes. Since there are two recombinant gametes, each has a proportion of 1/2 θ. • 1 - θ is the proportion of parental gametes. Each of the two parental gametes has a proportion of 1/2(1 - θ). Gametes: Parental: A B a b Recombinant: A b a B
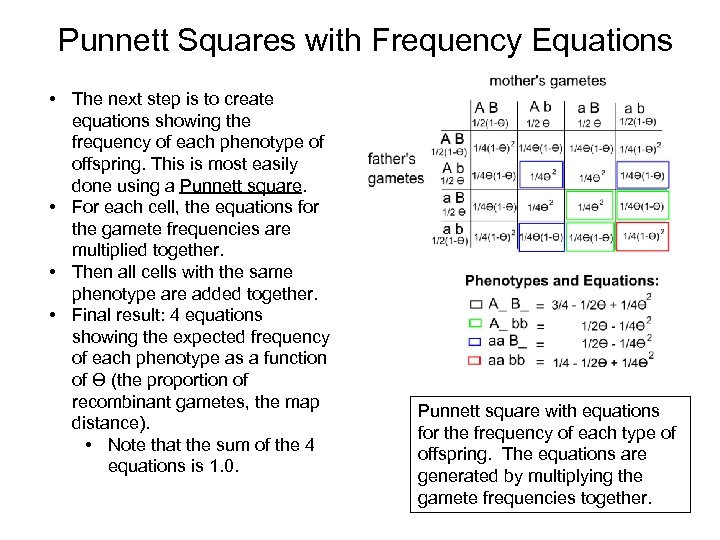
Punnett Squares with Frequency Equations • The next step is to create equations showing the frequency of each phenotype of offspring. This is most easily done using a Punnett square. • For each cell, the equations for the gamete frequencies are multiplied together. • Then all cells with the same phenotype are added together. • Final result: 4 equations showing the expected frequency of each phenotype as a function of Ɵ (the proportion of recombinant gametes, the map distance). • Note that the sum of the 4 equations is 1. 0. Punnett square with equations for the frequency of each type of offspring. The equations are generated by multiplying the gamete frequencies together.
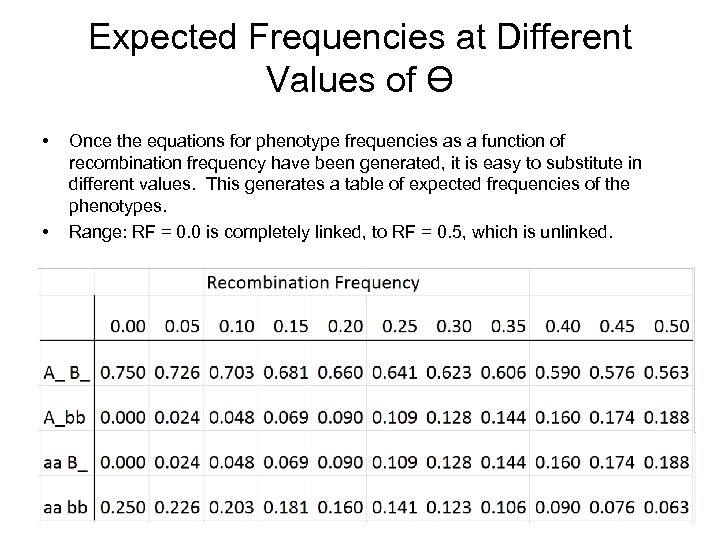
Expected Frequencies at Different Values of Ɵ • • Once the equations for phenotype frequencies as a function of recombination frequency have been generated, it is easy to substitute in different values. This generates a table of expected frequencies of the phenotypes. Range: RF = 0. 0 is completely linked, to RF = 0. 5, which is unlinked.
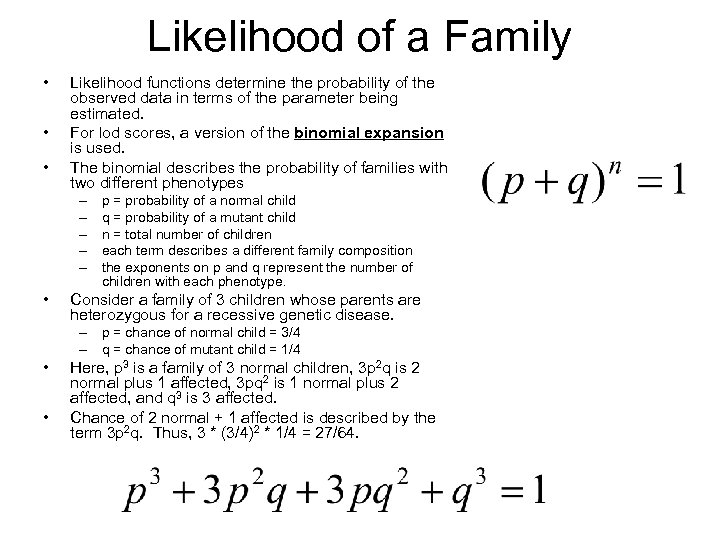
Likelihood of a Family • • • Likelihood functions determine the probability of the observed data in terms of the parameter being estimated. For lod scores, a version of the binomial expansion is used. The binomial describes the probability of families with two different phenotypes – – – • p = probability of a normal child q = probability of a mutant child n = total number of children each term describes a different family composition the exponents on p and q represent the number of children with each phenotype. Consider a family of 3 children whose parents are heterozygous for a recessive genetic disease. – p = chance of normal child = 3/4 – q = chance of mutant child = 1/4 • • Here, p 3 is a family of 3 normal children, 3 p 2 q is 2 normal plus 1 affected, 3 pq 2 is 1 normal plus 2 affected, and q 3 is 3 affected. Chance of 2 normal + 1 affected is described by the term 3 p 2 q. Thus, 3 * (3/4)2 * 1/4 = 27/64.
Multinomial Distribution • The multinomial distribution extends the binomial to more than two phenotypes. It is very simple: just add more components to each term. – For example, for 4 phenotypes, C p 2 q 1 r 3 s 1 (where C is some coefficient) describes the probability of a family of 7 children, where 2 of them have the “p” phenotype, 1 has the “q” phenotype, 3 have the “r” phenotype, and 1 has the “s” phenotype. • The coefficients in front of each term represent the number of possible families of the given composition. For the binomial we can calculate the coefficients using Pascal’s triangle (or a useful formula). • However, for LOD score mapping we don’t need to bother with the coefficients because they get divided out.
Likelihood Ratio • Using a spreadsheet, we first calculate the expected frequency of each type of offspring at different values of θ. • Then we use the data from actual families to calculate the likelihood of each family at each value of θ. • Then we take the likelihood ratio: divide the likelihood at each θ by the likelihood at θ = 0. 50 (i. e. unlinked). • Then we take the logarithm (base 10) of each likelihood.
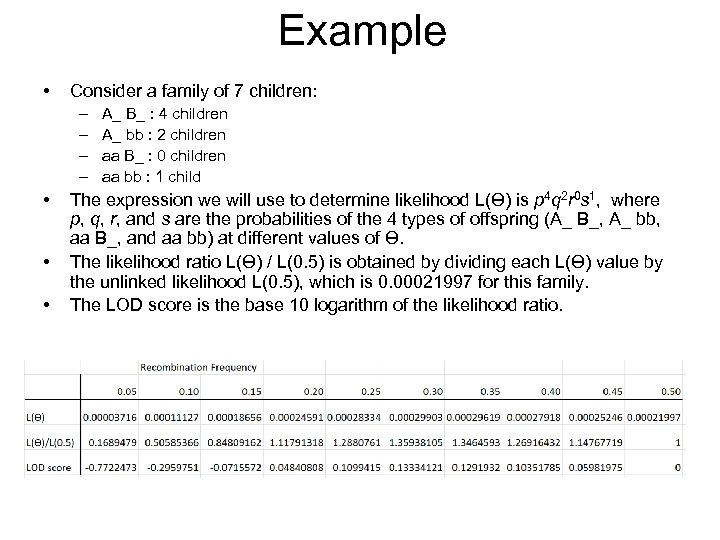
Example • Consider a family of 7 children: – – • • • A_ B_ : 4 children A_ bb : 2 children aa B_ : 0 children aa bb : 1 child The expression we will use to determine likelihood L(Ɵ) is p 4 q 2 r 0 s 1, where p, q, r, and s are the probabilities of the 4 types of offspring (A_ B_, A_ bb, aa B_, and aa bb) at different values of Ɵ. The likelihood ratio L(Ɵ) / L(0. 5) is obtained by dividing each L(Ɵ) value by the unlinked likelihood L(0. 5), which is 0. 00021997 for this family. The LOD score is the base 10 logarithm of the likelihood ratio.
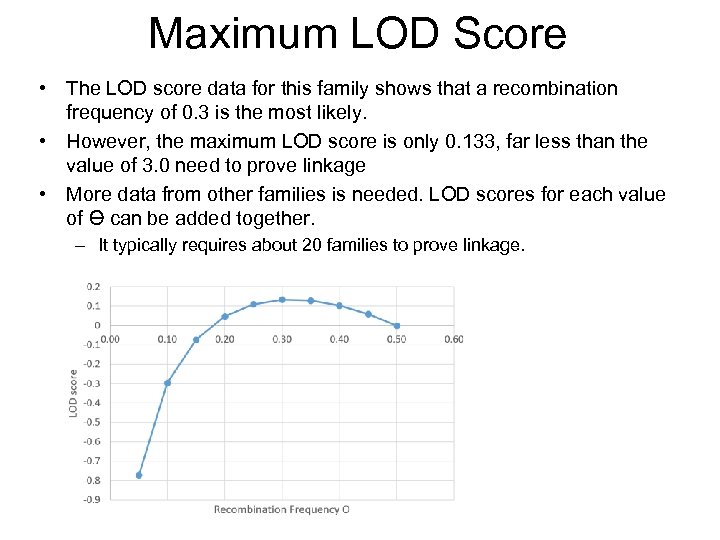
Maximum LOD Score • The LOD score data for this family shows that a recombination frequency of 0. 3 is the most likely. • However, the maximum LOD score is only 0. 133, far less than the value of 3. 0 need to prove linkage • More data from other families is needed. LOD scores for each value of Ɵ can be added together. – It typically requires about 20 families to prove linkage.
e188fc7eec78ff90229ef84d21047856.ppt