d775db79b2e1739f0afe8b3122a1ad50.ppt
- Количество слайдов: 33

Quick Overview of Bioinformatics NCBI Chuong Huynh NIH/NLM/NCBI New Delhi, India September 28, 2004 huynh@ncbi. nlm. nih. gov
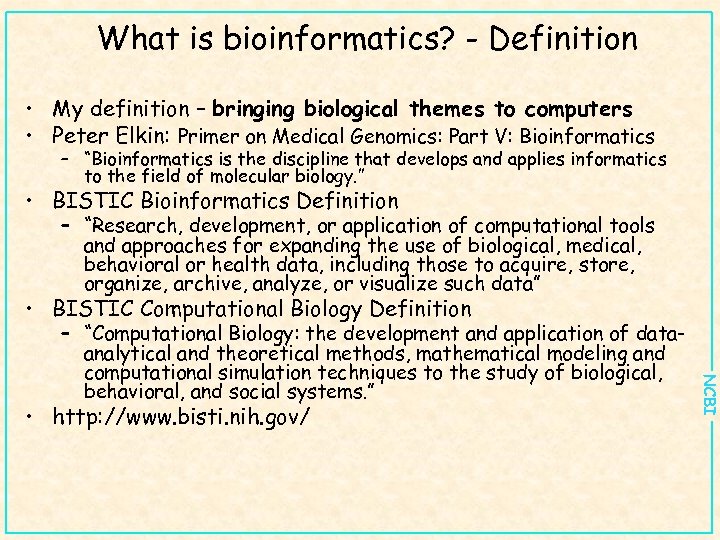
What is bioinformatics? - Definition • My definition – bringing biological themes to computers • Peter Elkin: Primer on Medical Genomics: Part V: Bioinformatics – “Bioinformatics is the discipline that develops and applies informatics to the field of molecular biology. ” • BISTIC Bioinformatics Definition – “Research, development, or application of computational tools and approaches for expanding the use of biological, medical, behavioral or health data, including those to acquire, store, organize, archive, analyze, or visualize such data” • BISTIC Computational Biology Definition • http: //www. bisti. nih. gov/ NCBI – “Computational Biology: the development and application of dataanalytical and theoretical methods, mathematical modeling and computational simulation techniques to the study of biological, behavioral, and social systems. ”
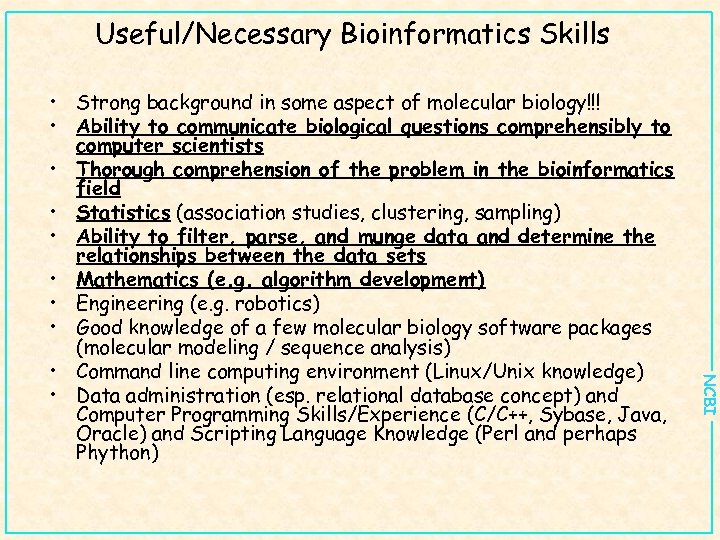
Useful/Necessary Bioinformatics Skills NCBI • Strong background in some aspect of molecular biology!!! • Ability to communicate biological questions comprehensibly to computer scientists • Thorough comprehension of the problem in the bioinformatics field • Statistics (association studies, clustering, sampling) • Ability to filter, parse, and munge data and determine the relationships between the data sets • Mathematics (e. g. algorithm development) • Engineering (e. g. robotics) • Good knowledge of a few molecular biology software packages (molecular modeling / sequence analysis) • Command line computing environment (Linux/Unix knowledge) • Data administration (esp. relational database concept) and Computer Programming Skills/Experience (C/C++, Sybase, Java, Oracle) and Scripting Language Knowledge (Perl and perhaps Phython)
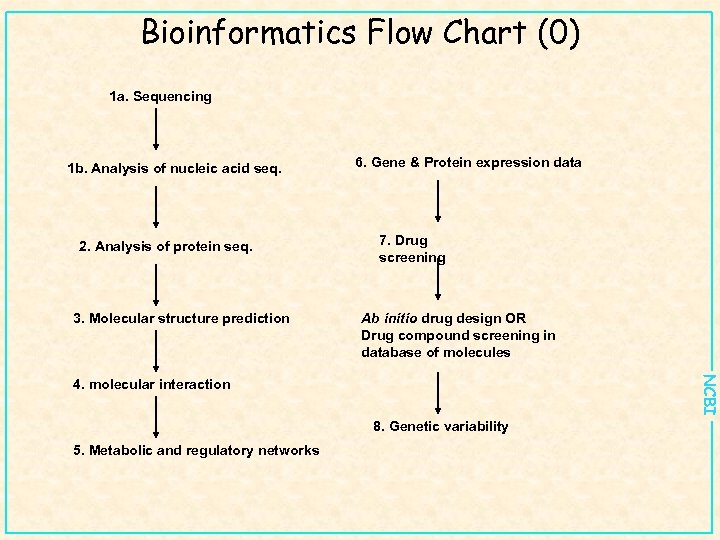
Bioinformatics Flow Chart (0) 1 a. Sequencing 1 b. Analysis of nucleic acid seq. 2. Analysis of protein seq. 3. Molecular structure prediction 6. Gene & Protein expression data 7. Drug screening Ab initio drug design OR Drug compound screening in database of molecules NCBI 4. molecular interaction 8. Genetic variability 5. Metabolic and regulatory networks
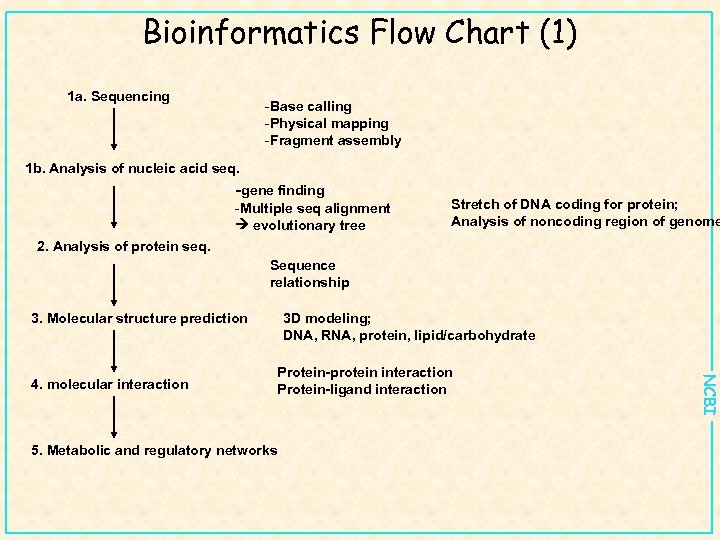
Bioinformatics Flow Chart (1) 1 a. Sequencing -Base calling -Physical mapping -Fragment assembly 1 b. Analysis of nucleic acid seq. -gene finding -Multiple seq alignment evolutionary tree Stretch of DNA coding for protein; Analysis of noncoding region of genome 2. Analysis of protein seq. Sequence relationship 3. Molecular structure prediction Protein-protein interaction Protein-ligand interaction 5. Metabolic and regulatory networks NCBI 4. molecular interaction 3 D modeling; DNA, RNA, protein, lipid/carbohydrate
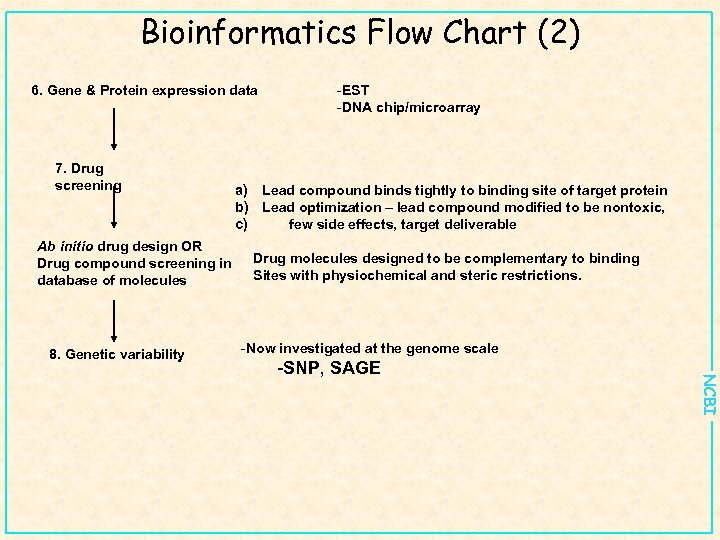
Bioinformatics Flow Chart (2) 6. Gene & Protein expression data 7. Drug screening Ab initio drug design OR Drug compound screening in database of molecules 8. Genetic variability -EST -DNA chip/microarray a) Lead compound binds tightly to binding site of target protein b) Lead optimization – lead compound modified to be nontoxic, c) few side effects, target deliverable Drug molecules designed to be complementary to binding Sites with physiochemical and steric restrictions. -Now investigated at the genome scale NCBI -SNP, SAGE
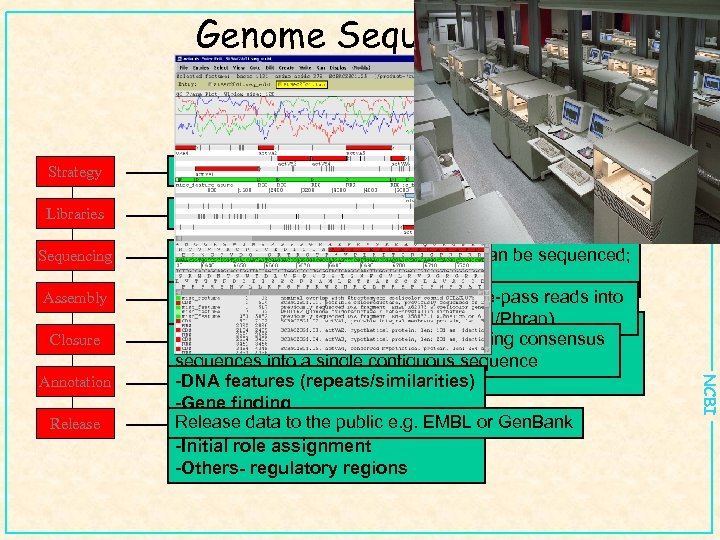
Genome Sequencing Strategy Clone by clone vs whole genome shotgun Libraries Subcloning; generate small insert libraries Sequencing Assembly Closure Release NCBI Annotation • Most genome will be sequenced and can be sequenced; few problem are unsolvable. Assembly: Process of taking raw single-pass reads into contiguous • Problem lies in understanding what you have: consensus sequence (Phred/Phrap) Closure: Process of ordering and merging consensus sequences into • Gene prediction/gene finding a single contiguous sequence • Annotation -DNA features (repeats/similarities) -Gene finding Release data to the public e. g. EMBL or Gen. Bank -Peptide features -Initial role assignment -Others- regulatory regions
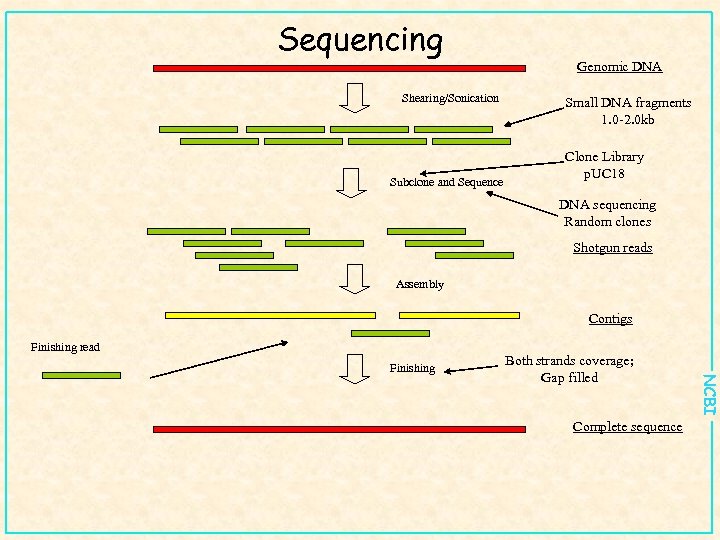
Sequencing Shearing/Sonication Subclone and Sequence Genomic DNA Small DNA fragments 1. 0 -2. 0 kb Clone Library p. UC 18 DNA sequencing Random clones Shotgun reads Assembly Contigs Finishing read Both strands coverage; Gap filled Complete sequence NCBI Finishing
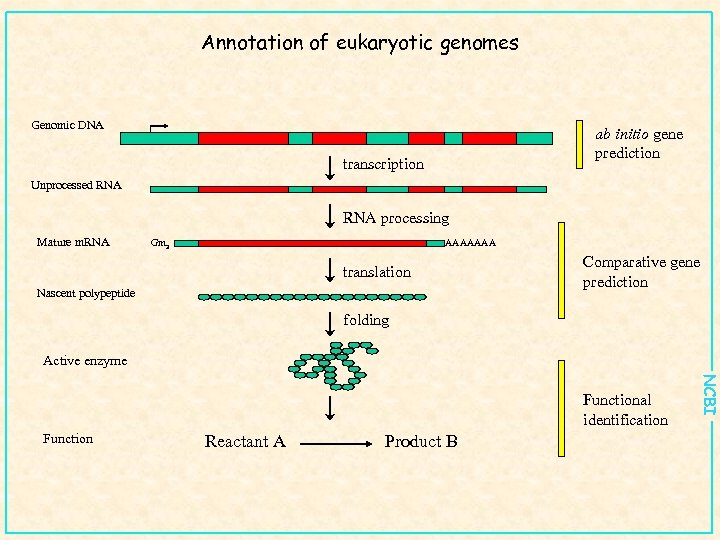
Annotation of eukaryotic genomes Genomic DNA ab initio gene prediction transcription Unprocessed RNA processing Mature m. RNA Gm 3 AAAAAAA translation Nascent polypeptide Comparative gene prediction folding Active enzyme Function Reactant A Product B NCBI Functional identification
Annotation • • NCBI Predict protein Extract ORFs Remove errors Compare with database of ‘known function proteins’ • Provide transitive annotations
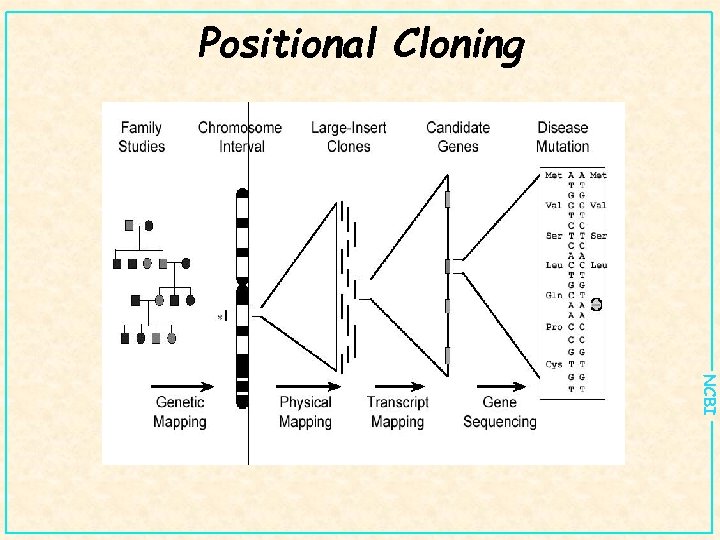
Positional Cloning NCBI
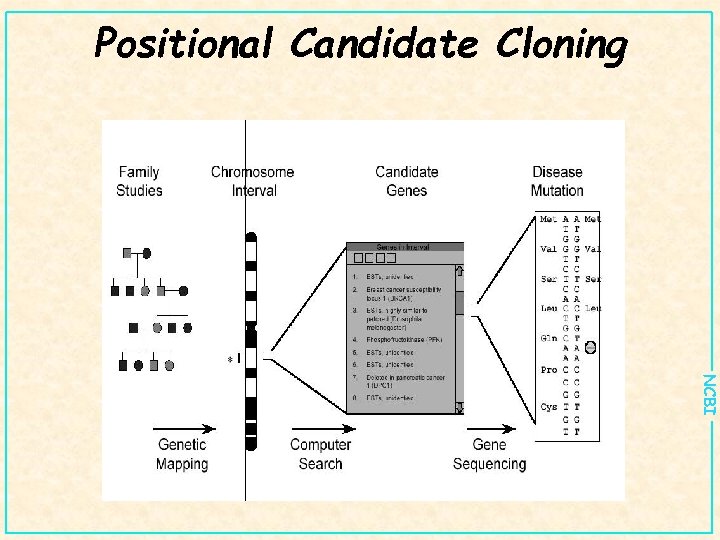
Positional Candidate Cloning NCBI

The new information is always partial • • • Complete Eukaryotic Genomes Ongoing Eukaryotic Prokaryotic Ongoing Published Even a complete genome is only partially understood NCBI
Why not use the genome sequence once its ‘ready’? • Finding exons • Expressed sequences are there in part and represent a very powerful key. NCBI – 30% overprediction – 20% not found at all – Comparison systems rely on EST sequences which themselves contain large error rates – Others are looking through partial data – Once the genome is done …when?
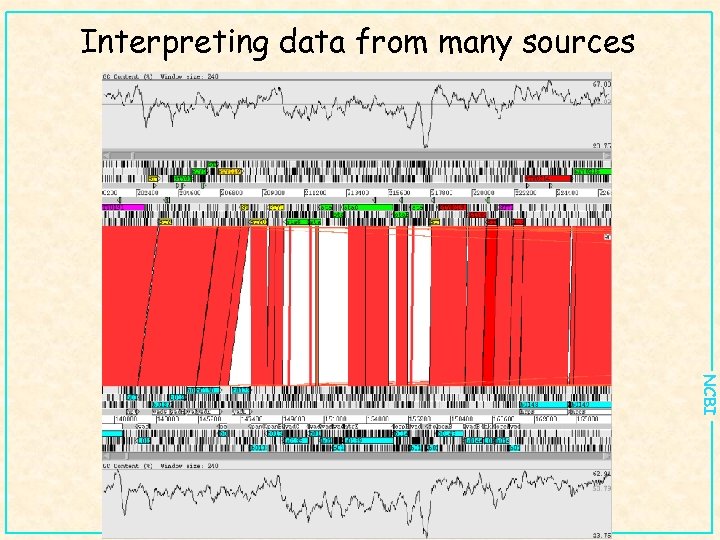
Interpreting data from many sources NCBI

Genomics and Tropical Diseases How Can Genomics Contribute to the Control of Tropical Diseases? Challenges and Opportunities The Role of Bioinformatics NCBI Strategic emphases for research http: //www. who. int/tdr/grants/strategic-emphases/default. htm WHO/TDR Genomics and World Health Report 2002
Why Pathogen Genomics? B. Bloom (1995) A microbial minimalist. Nature 378: 236 NCBI “The power and cost-effectiveness of modern genome sequencing technology mean that complete genome sequences of 25 of the major bacterial and parasitic pathogens could be available within five years. For about 100 million dollars (…), we could buy the sequence of every virulence determinant, every protein antigen and every drug target. ”
Genomics and Drug Development for Tropical Diseases: Challenges • Knowledge limitations – A large proportion of pathogen genes have unknown function – Heavy investment in genomics is done by the commercial sector and therefore not widely available • Emphasis and priorities NCBI – Genomes of non-pathogenic model organisms (S. cerevisiae, D. melanogaster, C. elegans, A. thaliana) – Genomes of pathogens that affect individuals in developed countries – Neglected diseases neglected pathogens

Doing Successful Science in the new millennium NCBI • Huge increase in available biological information • Classic paradigm of ‘molecular biology’ now is altering rapidly to genomics • Understanding of the new paradigms concerns more than ‘just bench biology’ • Discovery requires large scale systems and broad collaborations, Global problems • Funding comes in large amounts at group level, no longer a single laboratory or institution effort. • Accountable output
The Bigger Picture (Malaria) NCBI

Genomics Approach to Drug Development: Opportunities • Classical laboratory assays aim at targets in which mutation is lethal to the pathogen – Valuable targets can be missed NCBI • Sulphonamides: Inhibition of the p-aminobenzoic acid pathway not lethal for growth in laboratory but severely attenuate the capacity to cause disease
Genomics Approach to Drug Development: Opportunities • New approaches for the identification of gene products specifically involved in the disease process may uncover further drug targets • Pathogen genomics and data mining for the discovery of new drug targets NCBI – Signature tagged mutagenesis (STM) – Transposon site hybridization (Tra. SH)

Fosmidomycin • September 1999: a basic • 1 st semester 2001: Results of Phase I clinical trials NCBI science breakthrough (data mining through bioinformatics identify new targets for chemotherapy of malaria)

Fosmidomycin example - lesson • A lesson to take home: 1½ years from data mining and laboratory research to phase II, proof-ofprinciple clinical trials NCBI

Bioinformatics: Opportunities in Health Research and Development • New drug research and development – – – Identification of novel drug/vaccine targets Structural predictions Tapping into biodiversity Reconstruction of metabolic pathways Systems biology NCBI • Identification of vaccine candidates through analysis of surface antigens and epitopes

A Window of Opportunity for Disease Endemic Countries • Bioinformatics is an extremely important tool, with relevance to studying pathogenic organisms – Pathogens of interest to DECs already being sequenced (e. g. P. falciparum, T. cruzi, T. brucei, Leishmania sp. ) NCBI • Computational biology is ‘people-intensive’, less affected by infrastructure, economics, etc than other areas of biological research • ‘Critical mass’ issues less critical – a world-wide community is within reach

Relatively Modest Hardware Needs and Technical Support • Linux operating system permits use of the personal computer as a powerful workstation – Vast repository of public domain software for computational biology – EMB network nodes, FIOCRUZ (Brazil), SANBI (South Africa), CECALCULA (Venezuela), ICGEB (Trieste and New Delhi) NCBI • Individual accounts for remote access and data processing can be open at highperformance computer facilities and regional centers
Relatively Modest Hardware Needs and Technical Support • Powerful searches using public websites – NCBI, EMB nodes, Sanger Center, Expasy/Swiss. Prot, KEGG database • High-speed internet access is becoming more and more available in disease endemic countries through regional and international support, e. g. : NCBI – Asia-Pacific Advanced Network Consortium (APAN) http: //www. th. apan. net/ – MIMCom Malaria Research Resources http: //www. nlm. nih. gov/mimcom/about. html

International Training Course on Bioinformatics and Computational Biology Applied to Genome Studies (Train-the-trainers Workshop) May 21 -June 15, 2001 FIOCRUZ, Brazil NCBI TDR Regional Training Centers & Regional Training Courses on Bioinformatics Applied to Tropical Diseases • Africa – SANBI, Cape Town, South Africa • Course: Jan 20 -Feb 02, 2002; Mar 19 -Apr 4, 2003; Feb 215, 2004 (with NBN series) – Univ of Ibadan, Nigeria • Course: May 26 -Jun 07, 2003 • South America – USP, São Paulo, Brazil • Course: Feb 18 -March 02, 2002; July 17 -19, 2003; July 516, 2004; • Southeast Asia – ICGEB, New Delhi, India • Course: Apr 26 -May 09, 2002; Sep 22 -Oct 06, 2003; Sept 28 -Oct 11, 2004 – Mahidol University, Bangkok, Thailand • Course: Jul 09 -23, 2002; Sep 29 -Oct 10, 2003; July 26 Aug 6, 2004
Training Course on Bioinformatics and Functional Genomics Applied to Insect Vectors of Human Diseases At the Center for Bioinformatics and Applied Genomics (CBAG) and Center for Vector and Vector-Borne Diseases (CVVD), Faculty of Science, Mahidol University, Bangkok, Thailand January 17 -28, 2005 NCBI Training Course on Functional Genomics of Insect Vectors of Human Diseases African Center for Training in Functional Genomics of Insect Vectors of Human Diseases (AFRO VECTGEN) At the Malaria Research and Training Center (MRTC), Bamako, Mali Dec 1 -16, 2004

Beginning Bioinformatics Books NCBI • Baxevanis & Ouellette 2001. Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins 2 nd Edition. John Wiley Publishing. • Gibas & Jambeck 2001. Developing Bioinformatics Computer Skills. O’Reilly. • Bioinformatics: Genome Sequence Analysis Mount 2001 • Bioinformatics For Dummies – Claverie & Notredame 2003 • Bioinformatics and Functional Genomics Pesvner 2003 • Introduction to Bioinformatics – Lesk 2002 • Fundamental Concepts of Bioinformatics Krane & Raymer 2003 • Beginning Perl for Bioinformatics – Tisdall 2002 • Primer of Genome Science – Gibson & Muse 2002

Course Schedule Take out your course schedule. Comments and Suggestions NCBI

The Challenge What is expected of you? NCBI
d775db79b2e1739f0afe8b3122a1ad50.ppt