quantification of dna.ppt
- Количество слайдов: 20
 QUANTIFICATION OF DNA
QUANTIFICATION OF DNA
 Two methods are widely used to determine the concentration of DNA in solution. The most simple and accurate is the spectrophotometric method, but it has a relatively low sensitivity. If the total content of nucleic acids is small, the concentration of DNA can be determined from the intensity of their fluorescence in UV light after staining with ethidium bromide a. Electrophoretic Method For an approximate evaluation of the concentration of DNA, different amounts of marker nucleic acids are applied to the gel. The concentration of DNA in the sample is evaluated by comparing the fluorescence intensity of the sample and standard markers with known concentrations. It is important that the DNA samples are compared with the corresponding markers, since for the same amount of DNA the fluorescence intensity in UV light differs. B. Spot Method The concentration of nucleic acids in a solution can be determined by staining the solution with ethidium bromide by measuring the fluorescence intensity in UV light and comparing it with the fluorescence of markers of known concentration. As with the electrophoretic method, the markers must be nucleic acids of the corresponding type (i. e. , DNA)
Two methods are widely used to determine the concentration of DNA in solution. The most simple and accurate is the spectrophotometric method, but it has a relatively low sensitivity. If the total content of nucleic acids is small, the concentration of DNA can be determined from the intensity of their fluorescence in UV light after staining with ethidium bromide a. Electrophoretic Method For an approximate evaluation of the concentration of DNA, different amounts of marker nucleic acids are applied to the gel. The concentration of DNA in the sample is evaluated by comparing the fluorescence intensity of the sample and standard markers with known concentrations. It is important that the DNA samples are compared with the corresponding markers, since for the same amount of DNA the fluorescence intensity in UV light differs. B. Spot Method The concentration of nucleic acids in a solution can be determined by staining the solution with ethidium bromide by measuring the fluorescence intensity in UV light and comparing it with the fluorescence of markers of known concentration. As with the electrophoretic method, the markers must be nucleic acids of the corresponding type (i. e. , DNA)
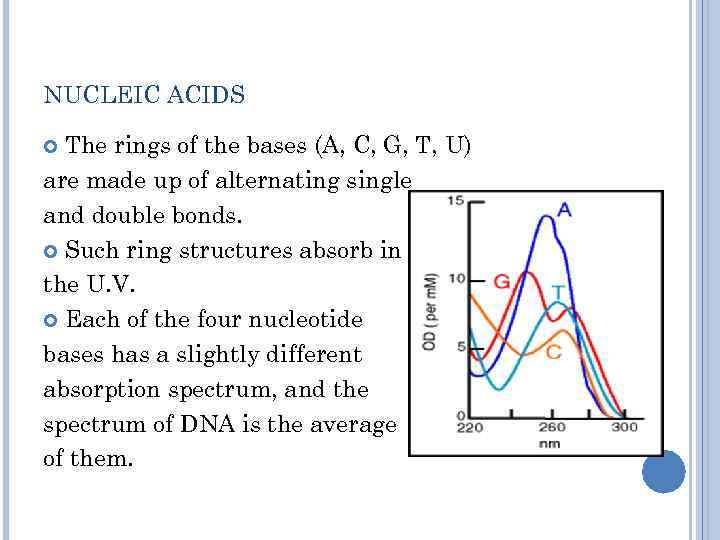 NUCLEIC ACIDS The rings of the bases (A, C, G, T, U) are made up of alternating single and double bonds. Such ring structures absorb in the U. V. Each of the four nucleotide bases has a slightly different absorption spectrum, and the spectrum of DNA is the average of them.
NUCLEIC ACIDS The rings of the bases (A, C, G, T, U) are made up of alternating single and double bonds. Such ring structures absorb in the U. V. Each of the four nucleotide bases has a slightly different absorption spectrum, and the spectrum of DNA is the average of them.
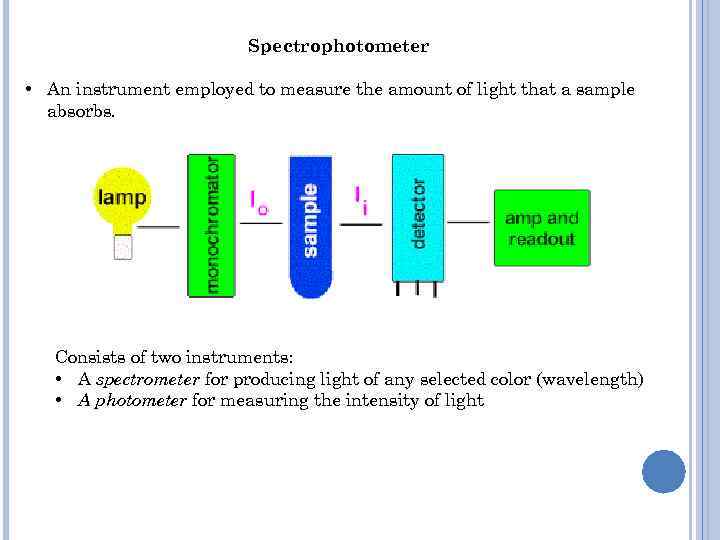 Spectrophotometer • An instrument employed to measure the amount of light that a sample absorbs. Consists of two instruments: • A spectrometer for producing light of any selected color (wavelength) • A photometer for measuring the intensity of light
Spectrophotometer • An instrument employed to measure the amount of light that a sample absorbs. Consists of two instruments: • A spectrometer for producing light of any selected color (wavelength) • A photometer for measuring the intensity of light
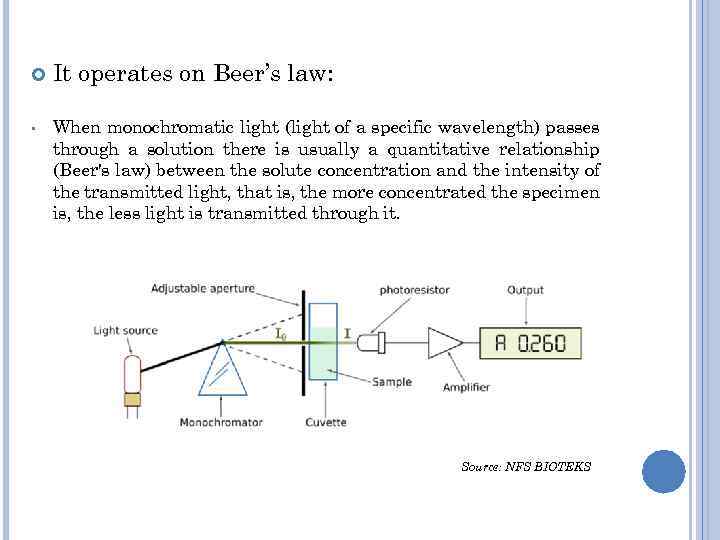 • It operates on Beer’s law: When monochromatic light (light of a specific wavelength) passes through a solution there is usually a quantitative relationship (Beer's law) between the solute concentration and the intensity of the transmitted light, that is, the more concentrated the specimen is, the less light is transmitted through it. Source: NFS BIOTEKS
• It operates on Beer’s law: When monochromatic light (light of a specific wavelength) passes through a solution there is usually a quantitative relationship (Beer's law) between the solute concentration and the intensity of the transmitted light, that is, the more concentrated the specimen is, the less light is transmitted through it. Source: NFS BIOTEKS
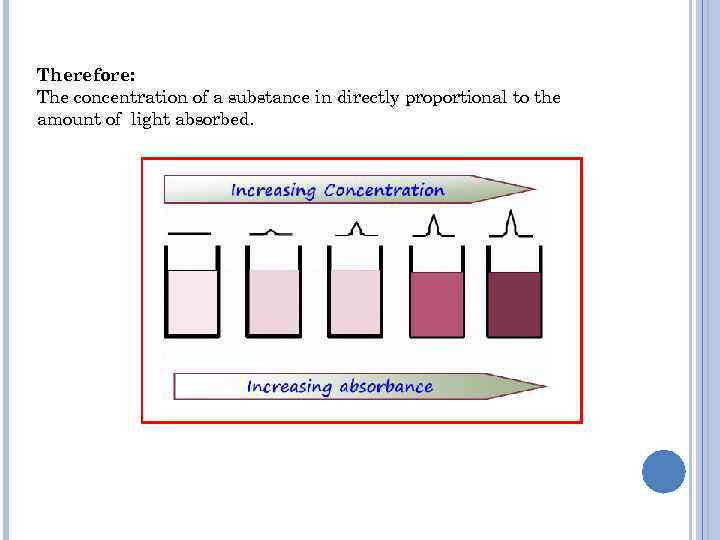 Therefore: The concentration of a substance in directly proportional to the amount of light absorbed.
Therefore: The concentration of a substance in directly proportional to the amount of light absorbed.
 DNA Analysis (After extraction) Nucleic acid (DNA/RNA) Qualitative analysis Quantitative analysis Size-Quality Purity- Yield Agarose gel electrophoresis Spectrophotometry
DNA Analysis (After extraction) Nucleic acid (DNA/RNA) Qualitative analysis Quantitative analysis Size-Quality Purity- Yield Agarose gel electrophoresis Spectrophotometry
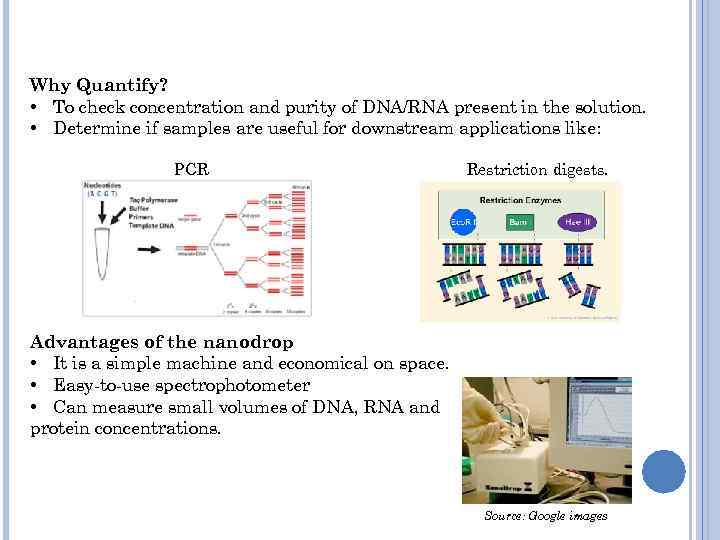 Why Quantify? • To check concentration and purity of DNA/RNA present in the solution. • Determine if samples are useful for downstream applications like: PCR Restriction digests. Advantages of the nanodrop • It is a simple machine and economical on space. • Easy-to-use spectrophotometer • Can measure small volumes of DNA, RNA and protein concentrations. Source: Google images
Why Quantify? • To check concentration and purity of DNA/RNA present in the solution. • Determine if samples are useful for downstream applications like: PCR Restriction digests. Advantages of the nanodrop • It is a simple machine and economical on space. • Easy-to-use spectrophotometer • Can measure small volumes of DNA, RNA and protein concentrations. Source: Google images
 Spectrophotometric measures • Measures DNA, RNA (A 260) and Proteins (A 280) concentrations and sample purity (260: 280). • Absorbance at 260 nm Nucleic acids absorb UV light at 260 nm due to the aromatic base moieties within their structure. Purines (thymine, cytosine and uracil) and pyrimidines (adenine and guanine) both have peak absorbances at 260 nm, thus making it the standard for quantitating nucleic acid samples. • Absorbance at 280 nm The 280 nm absorbance is measured where proteins and phenolic compounds have a strong absorbance. Similarly, the aromaticity of phenol groups of organic compounds absorbs strongly near 280 nm. • Absorbance at 230 nm Many organic compounds have strong absorbances at around 225 nm. In addition to phenol, TRIzol, and chaotropic salts, the peptide bonds in proteins absorb light between 200 and 230 nm.
Spectrophotometric measures • Measures DNA, RNA (A 260) and Proteins (A 280) concentrations and sample purity (260: 280). • Absorbance at 260 nm Nucleic acids absorb UV light at 260 nm due to the aromatic base moieties within their structure. Purines (thymine, cytosine and uracil) and pyrimidines (adenine and guanine) both have peak absorbances at 260 nm, thus making it the standard for quantitating nucleic acid samples. • Absorbance at 280 nm The 280 nm absorbance is measured where proteins and phenolic compounds have a strong absorbance. Similarly, the aromaticity of phenol groups of organic compounds absorbs strongly near 280 nm. • Absorbance at 230 nm Many organic compounds have strong absorbances at around 225 nm. In addition to phenol, TRIzol, and chaotropic salts, the peptide bonds in proteins absorb light between 200 and 230 nm.
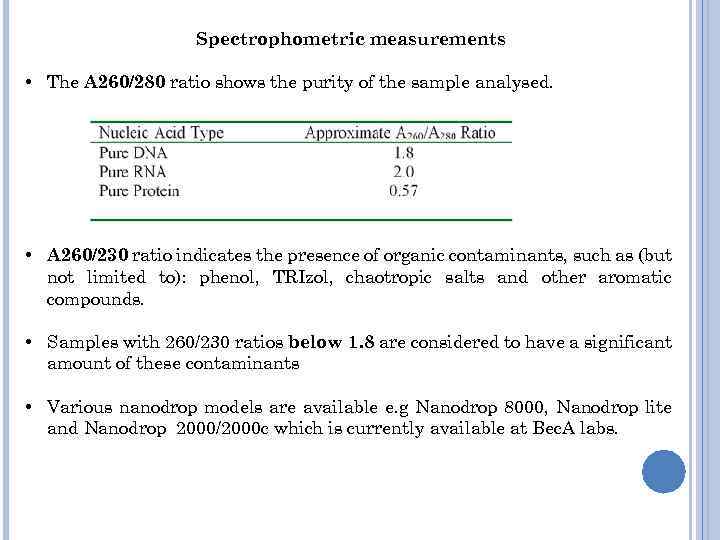 Spectrophometric measurements • The A 260/280 ratio shows the purity of the sample analysed. • A 260/230 ratio indicates the presence of organic contaminants, such as (but not limited to): phenol, TRIzol, chaotropic salts and other aromatic compounds. • Samples with 260/230 ratios below 1. 8 are considered to have a significant amount of these contaminants • Various nanodrop models are available e. g Nanodrop 8000, Nanodrop lite and Nanodrop 2000/2000 c which is currently available at Bec. A labs.
Spectrophometric measurements • The A 260/280 ratio shows the purity of the sample analysed. • A 260/230 ratio indicates the presence of organic contaminants, such as (but not limited to): phenol, TRIzol, chaotropic salts and other aromatic compounds. • Samples with 260/230 ratios below 1. 8 are considered to have a significant amount of these contaminants • Various nanodrop models are available e. g Nanodrop 8000, Nanodrop lite and Nanodrop 2000/2000 c which is currently available at Bec. A labs.
 Note that: • Because of the low concentrations, sometimes it is difficult to assess the purity of the samples (esp. RNA) by analyzing the A 260/280 and A 260/230 ratios. • Quantification cannot be assessed by the Nano. Drop because they are outside the lowest concentrations the Nano. Drop is designed to measure. • A more sensitive method such as an Agilent Biolanalyzer or Qubit analysis is recommended.
Note that: • Because of the low concentrations, sometimes it is difficult to assess the purity of the samples (esp. RNA) by analyzing the A 260/280 and A 260/230 ratios. • Quantification cannot be assessed by the Nano. Drop because they are outside the lowest concentrations the Nano. Drop is designed to measure. • A more sensitive method such as an Agilent Biolanalyzer or Qubit analysis is recommended.
 • Electrophoresis is a method whereby charged molecules in solution, chiefly proteins and nucleic acids, migrate in response to an electrical field. • Their rate of migration through the electrical field, depends on the strength of the field, on the net charge, size, and shape of the molecules, and also on the ionic strength, viscosity, and temperature of the medium in which the molecules are moving. • As an analytical tool, electrophoresis is simple, rapid and highly sensitive. • It can be used analytically to study the properties of a single charged species or mixtures of molecules. It can also be used preparatively as a separating technique
• Electrophoresis is a method whereby charged molecules in solution, chiefly proteins and nucleic acids, migrate in response to an electrical field. • Their rate of migration through the electrical field, depends on the strength of the field, on the net charge, size, and shape of the molecules, and also on the ionic strength, viscosity, and temperature of the medium in which the molecules are moving. • As an analytical tool, electrophoresis is simple, rapid and highly sensitive. • It can be used analytically to study the properties of a single charged species or mixtures of molecules. It can also be used preparatively as a separating technique
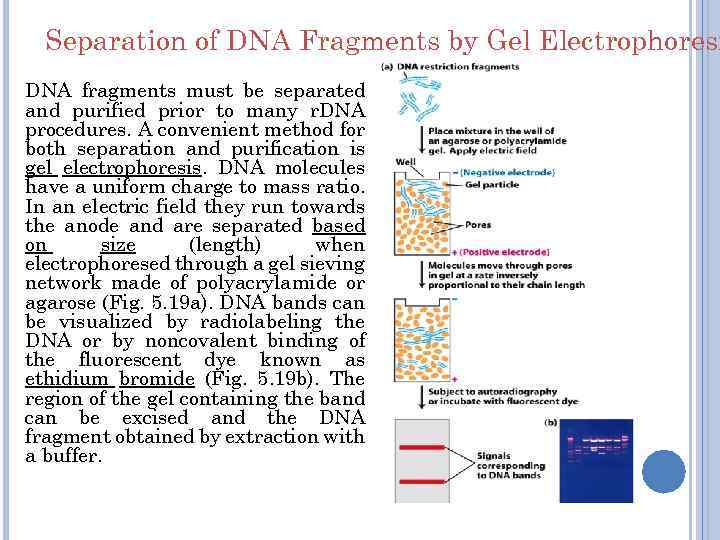 Separation of DNA Fragments by Gel Electrophoresi DNA fragments must be separated and purified prior to many r. DNA procedures. A convenient method for both separation and purification is gel electrophoresis. DNA molecules have a uniform charge to mass ratio. In an electric field they run towards the anode and are separated based on size (length) when electrophoresed through a gel sieving network made of polyacrylamide or agarose (Fig. 5. 19 a). DNA bands can be visualized by radiolabeling the DNA or by noncovalent binding of the fluorescent dye known as ethidium bromide (Fig. 5. 19 b). The region of the gel containing the band can be excised and the DNA fragment obtained by extraction with a buffer.
Separation of DNA Fragments by Gel Electrophoresi DNA fragments must be separated and purified prior to many r. DNA procedures. A convenient method for both separation and purification is gel electrophoresis. DNA molecules have a uniform charge to mass ratio. In an electric field they run towards the anode and are separated based on size (length) when electrophoresed through a gel sieving network made of polyacrylamide or agarose (Fig. 5. 19 a). DNA bands can be visualized by radiolabeling the DNA or by noncovalent binding of the fluorescent dye known as ethidium bromide (Fig. 5. 19 b). The region of the gel containing the band can be excised and the DNA fragment obtained by extraction with a buffer.
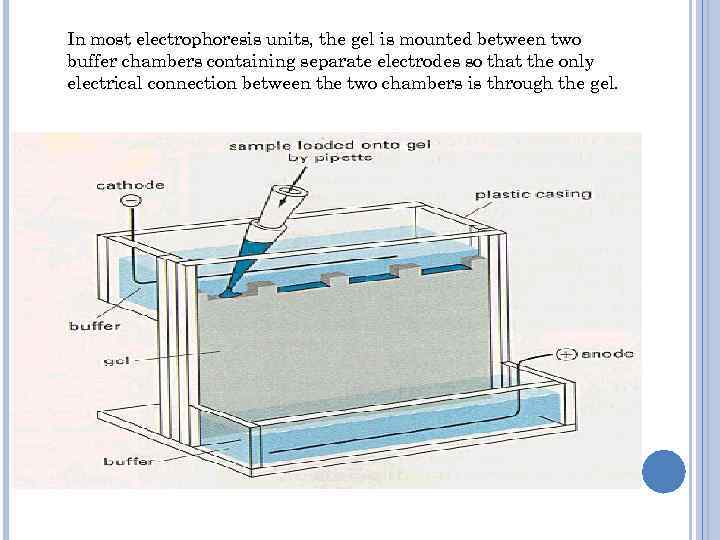 In most electrophoresis units, the gel is mounted between two buffer chambers containing separate electrodes so that the only electrical connection between the two chambers is through the gel.
In most electrophoresis units, the gel is mounted between two buffer chambers containing separate electrodes so that the only electrical connection between the two chambers is through the gel.
 Although agarose and polyacrylamide differ greatly in their physical and chemical structures, they both make porous gels. A porous gel acts as a sieve by retarding or, in some cases, by completely obstructing the movement of macromolecules while allowing smaller molecules to migrate freely. By preparing a gel with a restrictive pore size, the operator can take advantage of molecular size differences among proteins
Although agarose and polyacrylamide differ greatly in their physical and chemical structures, they both make porous gels. A porous gel acts as a sieve by retarding or, in some cases, by completely obstructing the movement of macromolecules while allowing smaller molecules to migrate freely. By preparing a gel with a restrictive pore size, the operator can take advantage of molecular size differences among proteins
 Fluorescent dyes are multicyclic molecules that absorb and emit fluorescent light at specific wavelengths. Examples are fluorescein and rhodamine derivatives. For sequencing applications, these molecules can be covalently attached to nucleotides.
Fluorescent dyes are multicyclic molecules that absorb and emit fluorescent light at specific wavelengths. Examples are fluorescein and rhodamine derivatives. For sequencing applications, these molecules can be covalently attached to nucleotides.
 In dye primer sequencing, the primer contains fluorescent dye–conjugated nucleotides, labeling the sequencing ladder at the 5′ ends of the chains. In dye terminator sequencing, the fluorescent dye
In dye primer sequencing, the primer contains fluorescent dye–conjugated nucleotides, labeling the sequencing ladder at the 5′ ends of the chains. In dye terminator sequencing, the fluorescent dye
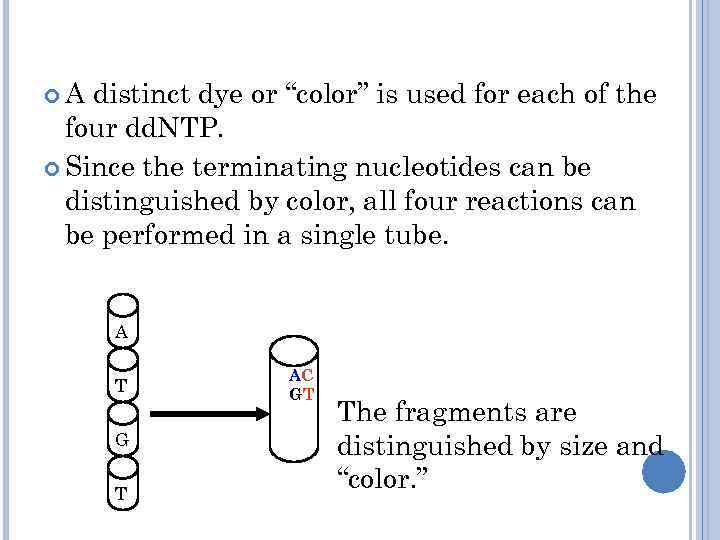 A distinct dye or “color” is used for each of the four dd. NTP. Since the terminating nucleotides can be distinguished by color, all four reactions can be performed in a single tube. A T G T AC GT The fragments are distinguished by size and “color. ”
A distinct dye or “color” is used for each of the four dd. NTP. Since the terminating nucleotides can be distinguished by color, all four reactions can be performed in a single tube. A T G T AC GT The fragments are distinguished by size and “color. ”
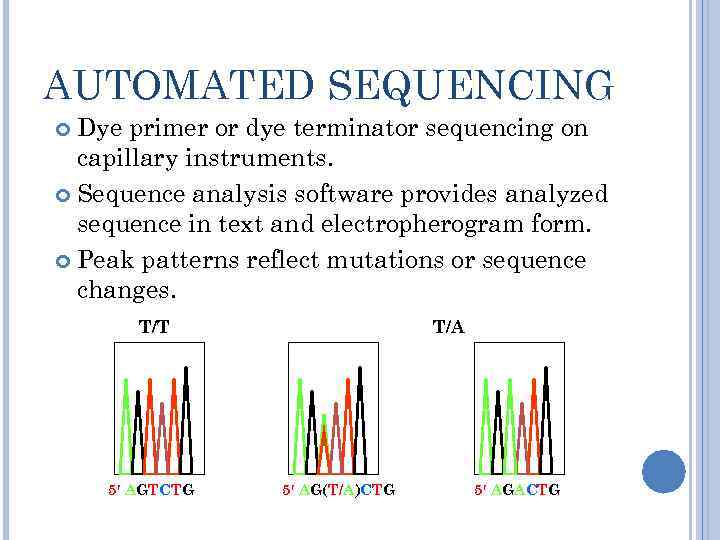 AUTOMATED SEQUENCING Dye primer or dye terminator sequencing on capillary instruments. Sequence analysis software provides analyzed sequence in text and electropherogram form. Peak patterns reflect mutations or sequence changes. T/T 5′ AGTCTG T/A 5′ AG(T/A)CTG 5′ AGACTG
AUTOMATED SEQUENCING Dye primer or dye terminator sequencing on capillary instruments. Sequence analysis software provides analyzed sequence in text and electropherogram form. Peak patterns reflect mutations or sequence changes. T/T 5′ AGTCTG T/A 5′ AG(T/A)CTG 5′ AGACTG
 THANK YOU
THANK YOU


